4BMW
 
 | | Crystal structure of the Streptomyces reticuli HbpS E78D, E81D double mutant | | Descriptor: | EXTRACELLULAR HAEM-BINDING PROTEIN | | Authors: | Wagener, S, Kursula, I, Wedderhoff, I, Groves, M.R, Ortiz de Orue Lucana, D. | | Deposit date: | 2013-05-11 | | Release date: | 2013-09-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Iron Binding at Specific Sites within the Octameric Hbps Protects Streptomycetes from Iron-Mediated Oxidative Stress.
Plos One, 8, 2013
|
|
5NFR
 
 | | Crystal structure of malate dehydrogenase from Plasmodium falciparum (PfMDH) | | Descriptor: | CITRIC ACID, Malate dehydrogenase | | Authors: | Lunev, S, Romero, A.R, Batista, F.A, Wrenger, C, Groves, M.R. | | Deposit date: | 2017-03-15 | | Release date: | 2018-01-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Oligomeric interfaces as a tool in drug discovery: Specific interference with activity of malate dehydrogenase of Plasmodium falciparum in vitro.
PLoS ONE, 13, 2018
|
|
7ZP2
 
 | | Crystal Structure of truncated aspartate transcarbamoylase from Plasmodium falciparum in complex with BDA-04 | | Descriptor: | Aspartate carbamoyltransferase, SODIUM ION, SULFATE ION, ... | | Authors: | Wang, C, Zhang, B, Groves, M.R. | | Deposit date: | 2022-04-26 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.292 Å) | | Cite: | Discovery of Small-Molecule Allosteric Inhibitors of Pf ATC as Antimalarials.
J.Am.Chem.Soc., 144, 2022
|
|
7ZST
 
 | | Crystal Structure of truncated aspartate transcarbamoylase from Plasmodium falciparum in complex with FLA-01 | | Descriptor: | 2-azanyl-~{N}-(2-methoxyethyl)-5-phenyl-thiophene-3-carboxamide, Aspartate carbamoyltransferase, GLYCEROL, ... | | Authors: | Wang, C, Zhang, B, Groves, M.R. | | Deposit date: | 2022-05-08 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of Small-Molecule Allosteric Inhibitors of Pf ATC as Antimalarials.
J.Am.Chem.Soc., 144, 2022
|
|
5OJE
 
 | | Endothiapepsin with Ligand VSK-B24 | | Descriptor: | (2~{S})-2-azanyl-3-(1~{H}-indol-3-yl)-~{N}-[2-(2,4,6-trimethylphenyl)ethyl]propanamide, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Gierse, R.M, Magari, F, Groves, M.R, Heine, A, Klebe, G, Hirsch, A. | | Deposit date: | 2017-07-21 | | Release date: | 2018-09-12 | | Last modified: | 2018-11-21 | | Method: | X-RAY DIFFRACTION (1.579 Å) | | Cite: | Design and Synthesis of Bioisosteres of Acylhydrazones as Stable Inhibitors of the Aspartic Protease Endothiapepsin.
ChemMedChem, 13, 2018
|
|
5FTP
 
 | | sulfur SAD phasing of Cdc23Nterm: data collection with a tailored X- ray beam size at 2.69 A wavelength (4.6 keV) | | Descriptor: | ANAPHASE-PROMOTING COMPLEX SUBUNIT 8 | | Authors: | Cianci, M, Groves, M.R, Barford, D, Schneider, T.R. | | Deposit date: | 2016-01-14 | | Release date: | 2016-03-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Data Collection with a Tailored X-Ray Beam Size at 2.69 A Wavelength (4.6 Kev): Sulfur Sad Phasing of Cdc23Nterm
Acta Crystallogr.,Sect.D, 72, 2016
|
|
5ILQ
 
 | | Crystal structure of truncated unliganded Aspartate Transcarbamoylase from Plasmodium falciparum | | Descriptor: | Aspartate carbamoyltransferase, GLYCEROL, SULFATE ION | | Authors: | Lunev, S, Bosch, S.S, Batista, F.D.A, Wrenger, C, Groves, M.R. | | Deposit date: | 2016-03-04 | | Release date: | 2016-06-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of truncated aspartate transcarbamoylase from Plasmodium falciparum.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5ILN
 
 | | Crystal structure of Aspartate Transcarbamoylase from Plasmodium falciparum (PfATC) with bound citrate | | Descriptor: | Aspartate carbamoyltransferase, CITRATE ANION, GLYCEROL, ... | | Authors: | Lunev, S, Bosch, S.S, Batista, F.D.A, Wrenger, C, Groves, M.R. | | Deposit date: | 2016-03-04 | | Release date: | 2017-03-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of truncated Aspartate Transcarbamoylase from Plasmodium falciparum
To be published
|
|
5MC7
 
 | |
5NKZ
 
 | | Crystal structure of H. polymorpha ubiquitin conjugating enzyme Pex4p in complex with soluble domain of Pex22p | | Descriptor: | Peroxin 22, Ubiquitin-conjugating enzyme E2-21 kDa | | Authors: | Danda, N, Lunev, S, Ali, A, Groves, M.R, Williams, C. | | Deposit date: | 2017-04-03 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural insights into K48-linked ubiquitin chain formation by the Pex4p-Pex22p complex.
Biochem. Biophys. Res. Commun., 496, 2018
|
|
3FPW
 
 | | Crystal Structure of HbpS with bound iron | | Descriptor: | Extracellular haem-binding protein, FE (III) ION, PHOSPHATE ION | | Authors: | Ortiz de Orue Lucana, D, Bogel, G, Zou, P, Groves, M.R. | | Deposit date: | 2009-01-06 | | Release date: | 2009-01-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The oligomeric assembly of the novel haem-degrading protein HbpS is essential for interaction with its cognate two-component sensor kinase
J.Mol.Biol., 386, 2009
|
|
3FPV
 
 | | Crystal Structure of HbpS | | Descriptor: | Extracellular haem-binding protein, FE (III) ION | | Authors: | Ortiz de Orue Lucana, D, Bogel, G, Zou, P, Groves, M.R. | | Deposit date: | 2009-01-06 | | Release date: | 2009-01-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The oligomeric assembly of the novel haem-degrading protein HbpS is essential for interaction with its cognate two-component sensor kinase
J.Mol.Biol., 386, 2009
|
|
1MHN
 
 | |
3L1P
 
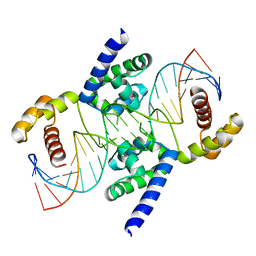 | | POU protein:DNA complex | | Descriptor: | DNA (5'-D(*AP*TP*CP*CP*AP*TP*TP*TP*GP*CP*CP*TP*TP*TP*CP*AP*AP*AP*TP*GP*TP*GP*G)-3'), DNA (5'-D(*TP*CP*CP*AP*CP*AP*TP*TP*TP*GP*AP*AP*AP*GP*GP*CP*AP*AP*AP*TP*GP*GP*A)-3'), POU domain, ... | | Authors: | Vahokoski, J, Groves, M.R, Pogenberg, V, Wilmanns, M. | | Deposit date: | 2009-12-14 | | Release date: | 2010-12-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A unique Oct4 interface is crucial for reprogramming to pluripotency
Nat.Cell Biol., 15, 2013
|
|
8B32
 
 | |
8B35
 
 | |
8B3C
 
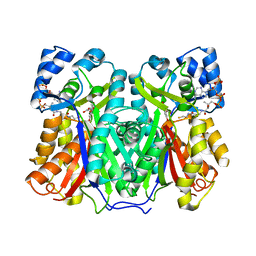 | | Chalcone synthase from Hordeum vulgare complexed with CoA and eriodictyol | | Descriptor: | (2S)-2-(3,4-DIHYDROXYPHENYL)-5,7-DIHYDROXY-2,3-DIHYDRO-4H-CHROMEN-4-ONE, COENZYME A, Chalcone synthase 2 | | Authors: | Zhang, L, Groves, M.R. | | Deposit date: | 2022-09-16 | | Release date: | 2023-09-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Engineering a Plant Polyketide Synthase for the Biosynthesis of Methylated Flavonoids.
J.Agric.Food Chem., 72, 2024
|
|
8C9T
 
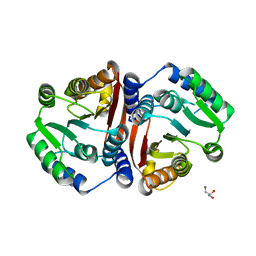 | | Catechol O-methyltransferase from Streptomyces avermitilis | | Descriptor: | GLYCEROL, Putative O-methyltransferase | | Authors: | Zhang, L, Groves, M.R. | | Deposit date: | 2023-01-23 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Characterization and Extended Substrate Scope Analysis of Two Mg 2+ -Dependent O-Methyltransferases from Bacteria.
Chembiochem, 24, 2023
|
|
8C9V
 
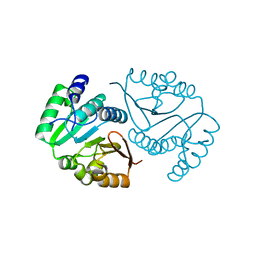 | | O-methyltransferase from Desulfuromonas acetoxidans | | Descriptor: | MAGNESIUM ION, O-methyltransferase, family 3 | | Authors: | Zhang, L, Groves, M.R. | | Deposit date: | 2023-01-23 | | Release date: | 2023-04-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Characterization and Extended Substrate Scope Analysis of Two Mg 2+ -Dependent O-Methyltransferases from Bacteria.
Chembiochem, 24, 2023
|
|
8C9S
 
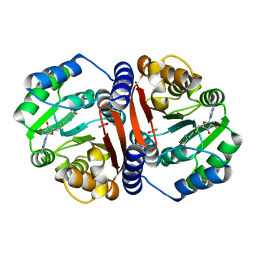 | |
1SMB
 
 | | Crystal Structure of Golgi-Associated PR-1 protein | | Descriptor: | 17kD fetal brain protein | | Authors: | Serrano, R.L, Kuhn, A, Hendricks, A, Helms, J.B, Sinning, I, Groves, M.R. | | Deposit date: | 2004-03-08 | | Release date: | 2004-09-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural analysis of the human Golgi-associated plant pathogenesis related protein GAPR-1 implicates dimerization as a regulatory mechanism
J.Mol.Biol., 339, 2004
|
|
7NT2
 
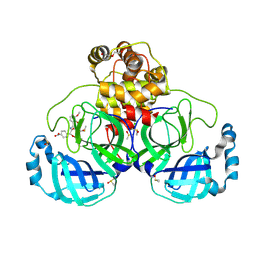 | | Crystal structure of SARS CoV2 main protease in complex with FSP006 | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Oerlemans, R, Eris, D, Wang, M, Sharpe, M, Domling, A, Groves, M.R. | | Deposit date: | 2021-03-08 | | Release date: | 2021-06-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.145 Å) | | Cite: | Combining High-Throughput Synthesis and High-Throughput Protein Crystallography for Accelerated Hit Identification.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7NT1
 
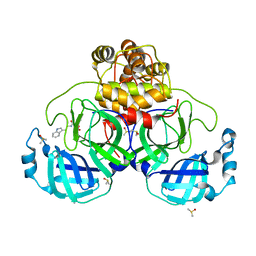 | | Crystal structure of SARS CoV2 main protease in complex with FSP007 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, [(2R)-1-[2-(1H-indol-3-yl)ethylamino]-1-oxidanylidene-butan-2-yl] prop-2-enoate | | Authors: | Oerlemans, R, Eris, D, Wang, M, Sharpe, M, Domling, A, Groves, M.R. | | Deposit date: | 2021-03-08 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Combining High-Throughput Synthesis and High-Throughput Protein Crystallography for Accelerated Hit Identification.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7NT3
 
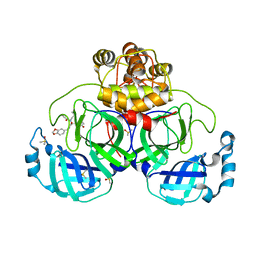 | | Crystal structure of SARS CoV2 main protease in complex with FSCU015 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ~{N}-[(1~{S})-2-(1,3-benzodioxol-5-ylmethylamino)-1-(3-hydroxyphenyl)-2-oxidanylidene-ethyl]-~{N}-propyl-prop-2-enamide | | Authors: | Oerlemans, R, Eris, D, Wang, M, Sharpe, M, Domling, A, Groves, M.R. | | Deposit date: | 2021-03-08 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.325 Å) | | Cite: | Combining High-Throughput Synthesis and High-Throughput Protein Crystallography for Accelerated Hit Identification.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7NTV
 
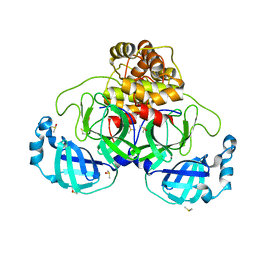 | | Crystal structure of SARS CoV2 main protease in complex with DN_EG_002 (modelled using PanDDA event map) | | Descriptor: | 2-acetamido-N-cyclopropyl-5-phenyl-thiophene-3-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Oerlemans, R, Eris, D, Wang, M, Sharpe, M, Domling, A, Groves, M.R. | | Deposit date: | 2021-03-10 | | Release date: | 2021-06-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.065 Å) | | Cite: | Combining High-Throughput Synthesis and High-Throughput Protein Crystallography for Accelerated Hit Identification.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
