2RP3
 
 | |
4FBV
 
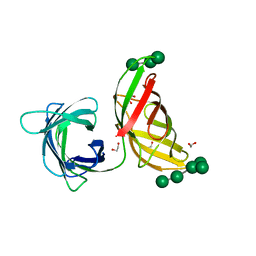 | | Crystal structure of the Myxococcus Xanthus hemagglutinin in complex with a3,a6-mannopentaose | | Descriptor: | 1,2-ETHANEDIOL, Myxobacterial hemagglutinin, alpha-D-mannopyranose, ... | | Authors: | Koharudin, L.M.I, Gronenborn, A.M. | | Deposit date: | 2012-05-23 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural Insights into the Anti-HIV Activity of the Oscillatoria agardhii Agglutinin Homolog Lectin Family.
J.Biol.Chem., 287, 2012
|
|
1ITI
 
 | | THE HIGH RESOLUTION THREE-DIMENSIONAL SOLUTION STRUCTURE OF HUMAN INTERLEUKIN-4 DETERMINED BY MULTI-DIMENSIONAL HETERONUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | Descriptor: | INTERLEUKIN-4 | | Authors: | Clore, G.M, Powers, B, Garrett, D.S, Gronenborn, A.M. | | Deposit date: | 1993-04-12 | | Release date: | 1993-07-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The high-resolution, three-dimensional solution structure of human interleukin-4 determined by multidimensional heteronuclear magnetic resonance spectroscopy.
Biochemistry, 32, 1993
|
|
1BBN
 
 | |
3GAT
 
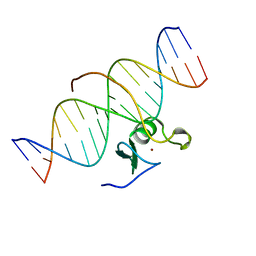 | | SOLUTION NMR STRUCTURE OF THE C-TERMINAL DOMAIN OF CHICKEN GATA-1 BOUND TO DNA, 34 STRUCTURES | | Descriptor: | DNA (5'-D(*AP*AP*TP*GP*TP*TP*TP*AP*TP*CP*TP*GP*CP*AP*AP*C)-3'), DNA (5'-D(*GP*TP*TP*GP*CP*AP*GP*AP*TP*AP*AP*AP*CP*AP*TP*T)-3'), ERYTHROID TRANSCRIPTION FACTOR GATA-1, ... | | Authors: | Clore, G.M, Tjandra, N, Starich, M, Omichinski, J.G, Gronenborn, A.M. | | Deposit date: | 1997-11-07 | | Release date: | 1998-01-28 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Use of dipolar 1H-15N and 1H-13C couplings in the structure determination of magnetically oriented macromolecules in solution.
Nat.Struct.Biol., 4, 1997
|
|
4Z8L
 
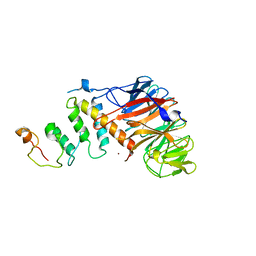 | | Crystal structure of DCAF1/SIV-MND VPX/MND SAMHD1 NTD ternary complex | | Descriptor: | Protein VPRBP, SAM domain and HD domain-containing protein, Vpx protein, ... | | Authors: | Koharudin, L.M, Wu, Y, Calero, G, Ahn, J, Gronenborn, A.M. | | Deposit date: | 2015-04-09 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis of Clade-specific Engagement of SAMHD1 (Sterile alpha Motif and Histidine/Aspartate-containing Protein 1) Restriction Factors by Lentiviral Viral Protein X (Vpx) Virulence Factors.
J.Biol.Chem., 290, 2015
|
|
1WJF
 
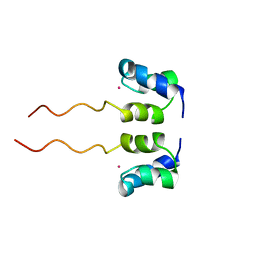 | | SOLUTION STRUCTURE OF H12C MUTANT OF THE N-TERMINAL ZN BINDING DOMAIN OF HIV-1 INTEGRASE COMPLEXED TO CADMIUM, NMR, 40 STRUCTURES | | Descriptor: | CADMIUM ION, HIV-1 INTEGRASE | | Authors: | Cai, M, Gronenborn, A.M, Clore, G.M. | | Deposit date: | 1998-06-11 | | Release date: | 1998-12-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the His12 --> Cys mutant of the N-terminal zinc binding domain of HIV-1 integrase complexed to cadmium.
Protein Sci., 7, 1998
|
|
3LHC
 
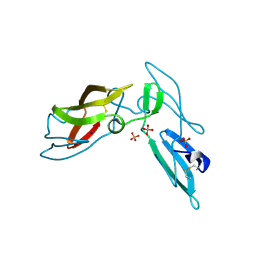 | | Crystal structure of cyanovirin-n swapping domain b mutant | | Descriptor: | Cyanovirin-N, PHOSPHATE ION, SODIUM ION | | Authors: | Matei, E, Zheng, A, Furey, W, Rose, J, Aiken, C, Gronenborn, A.M. | | Deposit date: | 2010-01-21 | | Release date: | 2010-02-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Anti-HIV activity of defective cyanovirin-N mutants is restored by dimerization.
J.Biol.Chem., 285, 2010
|
|
3IL8
 
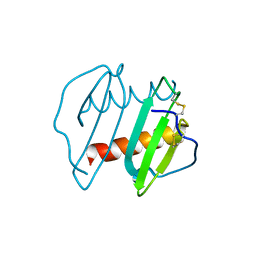 | | CRYSTAL STRUCTURE OF INTERLEUKIN 8: SYMBIOSIS OF NMR AND CRYSTALLOGRAPHY | | Descriptor: | INTERLEUKIN-8 | | Authors: | Baldwin, E.T, Weber, I.T, St Charles, R, Xuan, J.-C, Appella, E, Yamada, M, Matsushima, K, Edwards, B.F.P, Clore, G.M, Gronenborn, A.M, Wlodawer, A. | | Deposit date: | 1990-12-07 | | Release date: | 1992-10-15 | | Last modified: | 2012-02-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of interleukin 8: symbiosis of NMR and crystallography.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
3J34
 
 | | Structure of HIV-1 Capsid Protein by Cryo-EM | | Descriptor: | capsid protein | | Authors: | Zhao, G, Perilla, J.R, Yufenyuy, E, Meng, X, Chen, B, Ning, J, Ahn, J, Gronenborn, A.M, Schulten, K, Aiken, C, Zhang, P. | | Deposit date: | 2013-02-23 | | Release date: | 2013-05-29 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (8.6 Å) | | Cite: | Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics.
Nature, 497, 2013
|
|
1BBO
 
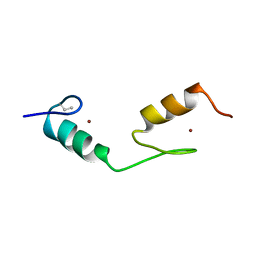 | |
1BBL
 
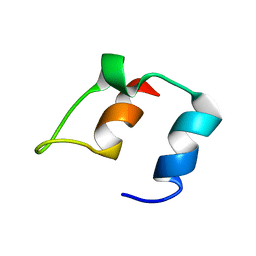 | |
1BAL
 
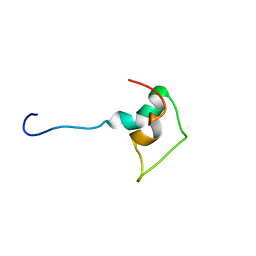 | |
4FBO
 
 | |
3OBL
 
 | |
1ATA
 
 | |
1ATB
 
 | |
1ATE
 
 | |
1ATD
 
 | |
4FBR
 
 | |
1L5I
 
 | | 30-CONFORMER NMR ENSEMBLE OF THE N-TERMINAL, DNA-BINDING DOMAIN OF THE REPLICATION INITIATION PROTEIN FROM A GEMINIVIRUS (TOMATO YELLOW LEAF CURL VIRUS-SARDINIA) | | Descriptor: | Rep protein | | Authors: | Campos-Olivas, R, Louis, J.M, Clerot, D, Gronenborn, B, Gronenborn, A.M. | | Deposit date: | 2002-03-07 | | Release date: | 2002-09-18 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The structure of a replication initiator unites diverse aspects of nucleic acid metabolism
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1L2M
 
 | | Minimized Average Structure of the N-terminal, DNA-binding domain of the replication initiation protein from a geminivirus (Tomato yellow leaf curl virus-Sardinia) | | Descriptor: | Rep protein | | Authors: | Campos-Olivas, R, Louis, J.M, Clerot, D, Gronenborn, B, Gronenborn, A.M. | | Deposit date: | 2002-02-22 | | Release date: | 2002-09-18 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The structure of a replication initiator unites diverse aspects of nucleic acid metabolism
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
2RMM
 
 | |
4QFY
 
 | | Crystal structure of the tetrameric dGTP/dCTP-bound SAMHD1 (RN206) mutant catalytic core | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ... | | Authors: | Koharudin, L.M.I, Wu, Y, DeLucia, M, Mehrens, J, Gronenborn, A.M, Ahn, J. | | Deposit date: | 2014-05-22 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis of Allosteric Activation of Sterile alpha Motif and Histidine-Aspartate Domain-containing Protein 1 (SAMHD1) by Nucleoside Triphosphates.
J.Biol.Chem., 289, 2014
|
|
4QFX
 
 | | Crystal structure of the tetrameric dGTP/dATP-bound SAMHD1 (RN206) mutant catalytic core | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ... | | Authors: | Koharudin, L.M.I, Wu, Y, DeLucia, M, Mehrens, J, Gronenborn, A.M, Ahn, J. | | Deposit date: | 2014-05-21 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of Allosteric Activation of Sterile alpha Motif and Histidine-Aspartate Domain-containing Protein 1 (SAMHD1) by Nucleoside Triphosphates.
J.Biol.Chem., 289, 2014
|
|
