5W8F
 
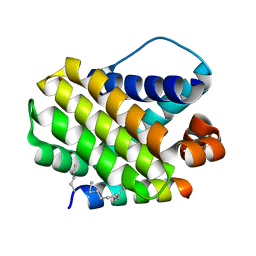 | | Crystal structure of human Mcl-1 in complex with modified Bim BH3 peptide SAH-MS1-14 | | Descriptor: | Induced myeloid leukemia cell differentiation protein Mcl-1, ZINC ION, modified Bim BH3 peptide SAH-MS1-14 | | Authors: | Rezaei Araghi, R, Jenson, J.M, Grant, R.A, Keating, A.E. | | Deposit date: | 2017-06-21 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Iterative optimization yields Mcl-1-targeting stapled peptides with selective cytotoxicity to Mcl-1-dependent cancer cells.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2QF0
 
 | |
2QAS
 
 | |
2QF3
 
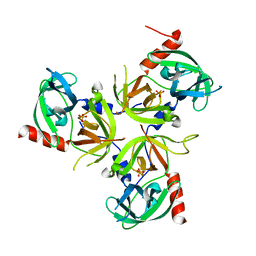 | | Structure of the delta PDZ truncation of the DegS protease | | Descriptor: | PHOSPHATE ION, Protease degS | | Authors: | Sohn, J, Grant, R.A, Sauer, R.T. | | Deposit date: | 2007-06-26 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Allosteric activation of DegS, a stress sensor PDZ protease.
Cell(Cambridge,Mass.), 131, 2007
|
|
8TWP
 
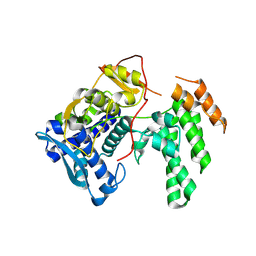 | | Influenza A virus (A/Aichi/2/1968(H3N2) nucleoprotein mutant - 2-7 deleted, R416A | | Descriptor: | Nucleoprotein | | Authors: | Yoon, J, Zhang, Y.M, Grant, R.A, Shoulders, M.D. | | Deposit date: | 2023-08-21 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The immune-evasive proline-283 substitution in influenza nucleoprotein increases aggregation propensity without altering the native structure.
Sci Adv, 10, 2024
|
|
8TWR
 
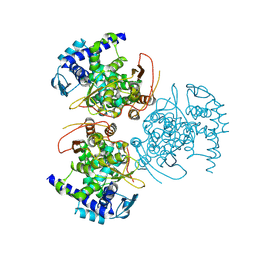 | | Influenza A virus (A/Aichi/2/1968(H3N2) nucleoprotein mutant - 2-7 deleted, P283S, R416A | | Descriptor: | Nucleoprotein, SODIUM ION | | Authors: | Yoon, J, Zhang, Y.M, Grant, R.A, Shoulders, M.D. | | Deposit date: | 2023-08-21 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | The immune-evasive proline-283 substitution in influenza nucleoprotein increases aggregation propensity without altering the native structure.
Sci Adv, 10, 2024
|
|
1PO2
 
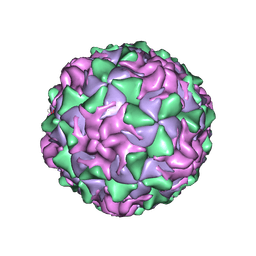 | | POLIOVIRUS (TYPE 1, MAHONEY) IN COMPLEX WITH R77975, AN INHIBITOR OF VIRAL REPLICATION | | Descriptor: | (METHYLPYRIDAZINE PIPERIDINE ETHYLOXYPHENYL)ETHYLACETATE, MYRISTIC ACID, POLIOVIRUS TYPE 1 MAHONEY | | Authors: | Hiremath, C.N, Filman, D.J, Grant, R.A, Hogle, J.M. | | Deposit date: | 1997-01-08 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Ligand-induced conformational changes in poliovirus-antiviral drug complexes.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
1PO1
 
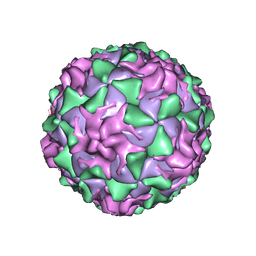 | | POLIOVIRUS (TYPE 1, MAHONEY) IN COMPLEX WITH R80633, AN INHIBITOR OF VIRAL REPLICATION | | Descriptor: | (METHYLPYRIDAZINE PIPERIDINE BUTYLOXYPHENYL)ETHYLACETATE, MYRISTIC ACID, POLIOVIRUS TYPE 1 MAHONEY | | Authors: | Hiremath, C.N, Filman, D.J, Grant, R.A, Hogle, J.M. | | Deposit date: | 1997-01-08 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Ligand-induced conformational changes in poliovirus-antiviral drug complexes.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
2RCE
 
 | |
2BVU
 
 | |
1PIV
 
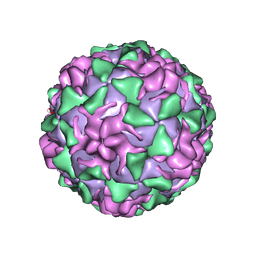 | | BINDING OF THE ANTIVIRAL DRUG WIN51711 TO THE SABIN STRAIN OF TYPE 3 POLIOVIRUS: STRUCTURAL COMPARISON WITH DRUG BINDING IN RHINOVIRUS 14 | | Descriptor: | 5-(7-(4-(4,5-DIHYDRO-2-OXAZOLYL)PHENOXY)HEPTYL)-3-METHYL ISOXAZOLE, MYRISTIC ACID, POLIOVIRUS TYPE 3 (SUBUNIT VP1), ... | | Authors: | Hiremath, C.N, Grant, R.A, Filman, D.J, Hogle, J.M. | | Deposit date: | 1995-02-02 | | Release date: | 1995-06-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Binding of the antiviral drug WIN51711 to the sabin strain of type 3 poliovirus: structural comparison with drug binding in rhinovirus 14.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
2QGR
 
 | |
6X0A
 
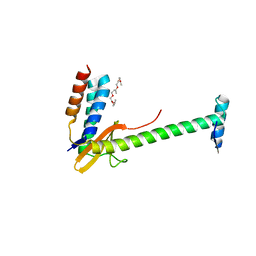 | | X-ray structure of a chimeric ParDE toxin-antitoxin complex from Mesorhizobium opportunistum | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, Plasmid stabilization system, Putative addiction module antidote protein, ... | | Authors: | Lite, T.L, Grant, R.A, Laub, M.T. | | Deposit date: | 2020-05-15 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Uncovering the basis of protein-protein interaction specificity with a combinatorially complete library.
Elife, 9, 2020
|
|
1YFN
 
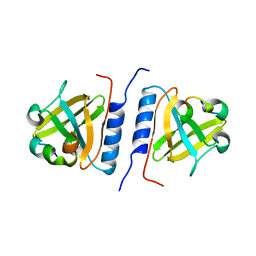 | | Versatile modes of peptide recognition by the AAA+ adaptor protein SspB- the crystal structure of a SspB-RseA complex | | Descriptor: | Sigma-E factor negative regulatory protein, Stringent starvation protein B | | Authors: | Levchenko, I, Grant, R.A, Flynn, J.M, Sauer, R.T, Baker, T.A. | | Deposit date: | 2005-01-03 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Versatile modes of peptide recognition by the AAA+ adaptor protein SspB
Nat.Struct.Mol.Biol., 12, 2005
|
|
1ZSZ
 
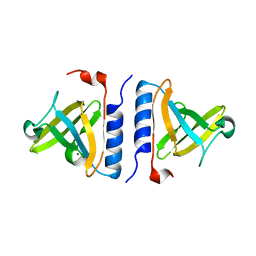 | | Crystal structure of a computationally designed SspB heterodimer | | Descriptor: | MAGNESIUM ION, Stringent starvation protein B homolog | | Authors: | Bolon, D.N, Grant, R.A, Baker, T.A, Sauer, R.T. | | Deposit date: | 2005-05-25 | | Release date: | 2005-08-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Specificity versus stability in computational protein design.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1OU8
 
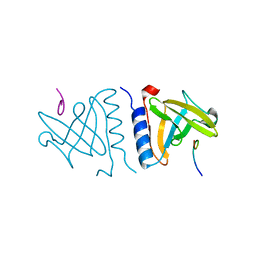 | | structure of an AAA+ protease delivery protein in complex with a peptide degradation tag | | Descriptor: | MAGNESIUM ION, Stringent starvation protein B homolog, synthetic ssrA peptide | | Authors: | Levchenko, I, Grant, R.A, Wah, D.A, Sauer, R.T, Baker, T.A. | | Deposit date: | 2003-03-24 | | Release date: | 2003-09-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of a delivery protein for an AAA+ protease in complex with a peptide degradation tag
Mol.Cell, 12, 2003
|
|
1OU9
 
 | | Structure of SspB, a AAA+ protease delivery protein | | Descriptor: | CALCIUM ION, Stringent starvation protein B homolog | | Authors: | Levchenko, I, Grant, R.A, Wah, D.A, Sauer, R.T, Baker, T.A. | | Deposit date: | 2003-03-24 | | Release date: | 2003-09-23 | | Last modified: | 2014-04-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a delivery protein for an AAA+ protease in complex with a peptide degradation tag
Mol.Cell, 12, 2003
|
|
1OUL
 
 | | Structure of the AAA+ protease delivery protein SspB | | Descriptor: | Stringent starvation protein B homolog | | Authors: | Levchenko, I, Grant, R.A, Wah, D.A, Sauer, R.T, Baker, T.A. | | Deposit date: | 2003-03-24 | | Release date: | 2003-09-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a delivery protein for an AAA+ protease in complex with a peptide degradation tag
Mol.Cell, 12, 2003
|
|
1U9P
 
 | | Permuted single-chain Arc | | Descriptor: | pArc | | Authors: | Tabtiang, R.K, Cezairliyan, B.O, Grant, R.A, Cochrane, J.C, Sauer, R.T. | | Deposit date: | 2004-08-10 | | Release date: | 2005-02-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Consolidating critical binding determinants by noncyclic rearrangement of protein secondary structure
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
4RQZ
 
 | |
4RR0
 
 | |
4RR1
 
 | |
1G2D
 
 | | STRUCTURE OF A CYS2HIS2 ZINC FINGER/TATA BOX COMPLEX (CLONE #2) | | Descriptor: | 5'-D(*GP*AP*CP*GP*CP*TP*AP*TP*AP*AP*AP*AP*GP*GP*AP*G)-3', 5'-D(*TP*CP*CP*TP*TP*TP*TP*AP*TP*AP*GP*CP*GP*TP*CP*C)-3', TATA BOX ZINC FINGER PROTEIN, ... | | Authors: | Wolfe, S.A, Grant, R.A, Elrod-Erickson, M, Pabo, C.O. | | Deposit date: | 2000-10-18 | | Release date: | 2001-09-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Beyond the "recognition code": structures of two Cys2His2 zinc finger/TATA box complexes.
Structure, 9, 2001
|
|
1TWB
 
 | | SspB disulfide crosslinked to an ssrA degradation tag | | Descriptor: | Stringent starvation protein B homolog, ssrA peptide | | Authors: | Bolon, D.N, Grant, R.A, Baker, T.A, Sauer, R.T. | | Deposit date: | 2004-06-30 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Nucleotide-Dependent Substrate Handoff from the SspB Adaptor to the AAA+ ClpXP Protease.
Mol.Cell, 16, 2004
|
|
6VWP
 
 | | Crystal structure of E. coli guanosine kinase in complex with ppGpp | | Descriptor: | GUANOSINE, GUANOSINE-5',3'-TETRAPHOSPHATE, Inosine-guanosine kinase, ... | | Authors: | Wang, B, Grant, R.A, Laub, M.T. | | Deposit date: | 2020-02-20 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | ppGpp Coordinates Nucleotide and Amino-Acid Synthesis in E. coli During Starvation.
Mol.Cell, 80, 2020
|
|
