1W2M
 
 | | Ca-substituted form of E. coli aminopeptidase P | | Descriptor: | CALCIUM ION, CHLORIDE ION, ISOPROPYL ALCOHOL, ... | | Authors: | Graham, S.C, Bond, C.S, Freeman, H.C, Guss, J.M. | | Deposit date: | 2004-07-07 | | Release date: | 2005-09-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Functional Implications of Metal Ion Selection in Aminopeptidase P, a Metalloprotease with a Dinuclear Metal Center.
Biochemistry, 44, 2005
|
|
2BWW
 
 | | His350Ala Escherichia coli Aminopeptidase P | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, AMINOPEPTIDASE P, CITRATE ANION, ... | | Authors: | Graham, S.C, Guss, J.M. | | Deposit date: | 2005-07-19 | | Release date: | 2006-01-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Kinetic and Crystallographic Analysis of Mutant Escherichia Coli Aminopeptidase P: Insights Into Substrate Recognition and the Mechanism of Catalysis.
Biochemistry, 45, 2006
|
|
2BH3
 
 | | Zn substituted E. coli Aminopeptidase P in complex with product | | Descriptor: | CITRATE ANION, LEUCINE, MAGNESIUM ION, ... | | Authors: | Graham, S.C, Bond, C.S, Freeman, H.C, Guss, J.M. | | Deposit date: | 2005-01-07 | | Release date: | 2005-09-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Functional Implications of Metal Ion Selection in Aminopeptidase P, a Metalloprotease with a Dinuclear Metal Center.
Biochemistry, 44, 2005
|
|
1WBQ
 
 | | ZnMg substituted aminopeptidase P from E. coli | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, XAA-PRO AMINOPEPTIDASE, ... | | Authors: | Graham, S.C, Bond, C.S, Freeman, H.C, Guss, J.M. | | Deposit date: | 2004-11-05 | | Release date: | 2005-09-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Functional Implications of Metal Ion Selection in Aminopeptidase P, a Metalloprotease with a Dinuclear Metal Center.
Biochemistry, 44, 2005
|
|
2UWI
 
 | | Structure of CrmE, a poxvirus TNF receptor | | Descriptor: | CRME PROTEIN | | Authors: | Graham, S.C, Bahar, M.W, Abrescia, N.G, Smith, G.L, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2007-03-22 | | Release date: | 2007-07-10 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Crme, a Virus-Encoded Tumour Necrosis Factor Receptor.
J.Mol.Biol., 372, 2007
|
|
2V3Z
 
 | |
2VVW
 
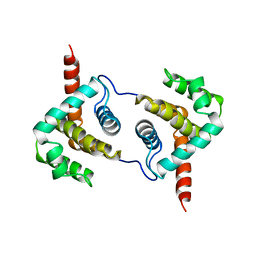 | | Structure of Vaccinia virus protein A52 | | Descriptor: | PROTEIN A52 | | Authors: | Graham, S.C, Bahar, M.W, Cooray, S, Chen, R.A.-J, Whalen, D.M, Abrescia, N.G.A, Alderton, D, Owens, R.J, Stuart, D.I, Smith, G.L, Grimes, J.M. | | Deposit date: | 2008-06-12 | | Release date: | 2008-08-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Vaccinia Virus Proteins A52 and B14 Share a Bcl-2-Like Fold But Have Evolved to Inhibit NF-kappaB Rather Than Apoptosis
Plos Pathog., 4, 2008
|
|
2VVY
 
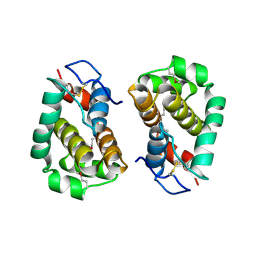 | | Structure of Vaccinia virus protein B14 | | Descriptor: | PROTEIN B15 | | Authors: | Graham, S.C, Bahar, M.W, Cooray, S, Chen, R.A.-J, Whalen, D.M, Abrescia, N.G.A, Alderton, D, Owens, R.J, Stuart, D.I, Smith, G.L, Grimes, J.M. | | Deposit date: | 2008-06-12 | | Release date: | 2008-08-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | Vaccinia Virus Proteins A52 and B14 Share a Bcl-2-Like Fold But Have Evolved to Inhibit NF-kappaB Rather Than Apoptosis
Plos Pathog., 4, 2008
|
|
2V3X
 
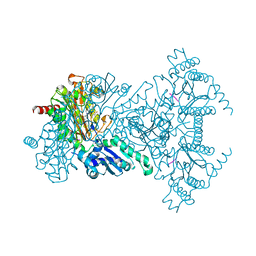 | |
2VVX
 
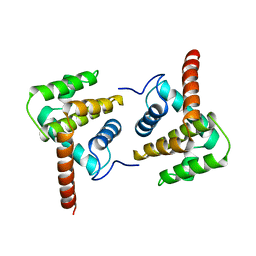 | | Structure of Vaccinia virus protein A52 | | Descriptor: | PROTEIN A52 | | Authors: | Graham, S.C, Bahar, M.W, Cooray, S, Chen, R.A.-J, Whalen, D.M, Abrescia, N.G.A, Alderton, D, Owens, R.J, Stuart, D.I, Smith, G.L, Grimes, J.M. | | Deposit date: | 2008-06-12 | | Release date: | 2008-08-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.746 Å) | | Cite: | Vaccinia Virus Proteins A52 and B14 Share a Bcl-2-Like Fold But Have Evolved to Inhibit NF-kappaB Rather Than Apoptosis
Plos Pathog., 4, 2008
|
|
2V3Y
 
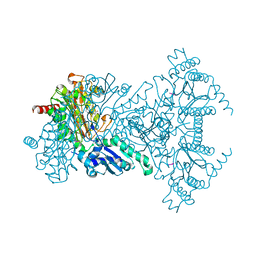 | |
2W2R
 
 | | Structure of the vesicular stomatitis virus matrix protein | | Descriptor: | MATRIX PROTEIN | | Authors: | Graham, S.C, Assenberg, R, Delmas, O, Verma, A, Gholami, A, Talbi, C, Owens, R.J, Stuart, D.I, Grimes, J.M, Bourhy, H. | | Deposit date: | 2008-11-03 | | Release date: | 2009-01-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Rhabdovirus Matrix Protein Structures Reveal a Novel Mode of Self-Association.
Plos Pathog., 4, 2008
|
|
7PHY
 
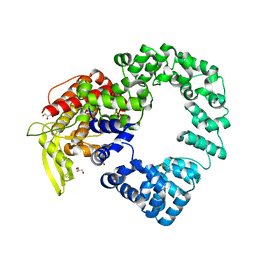 | | Vaccinia virus E2 | | Descriptor: | GLYCEROL, Protein E2 | | Authors: | Gao, W.N.D, Gao, C, Graham, S.C. | | Deposit date: | 2021-08-19 | | Release date: | 2021-09-01 | | Last modified: | 2022-02-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of vaccinia virus protein E2 and perspectives on the prediction of novel viral protein folds.
J.Gen.Virol., 103, 2022
|
|
7BNY
 
 | | Structure of 2A protein from encephalomyocarditis virus (EMCV) | | Descriptor: | Genome polyprotein, SULFATE ION | | Authors: | Hill, C.H, Napthine, S, Pekarek, L, Kibe, A, Firth, A.E, Graham, S.C, Caliskan, N, Brierley, I. | | Deposit date: | 2021-01-22 | | Release date: | 2021-12-08 | | Last modified: | 2022-02-02 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural and molecular basis for Cardiovirus 2A protein as a viral gene expression switch.
Nat Commun, 12, 2021
|
|
3ZYL
 
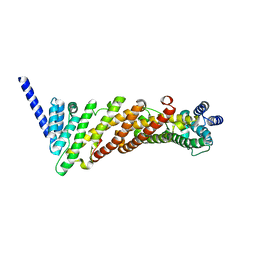 | | Structure of a truncated CALM (PICALM) ANTH domain | | Descriptor: | PHOSPHATIDYLINOSITOL-BINDING CLATHRIN ASSEMBLY PROTEIN | | Authors: | Miller, S.E, Sahlender, D.A, Graham, S.C, Honing, S, Robinson, M.S, Peden, A.A, Owen, D.J. | | Deposit date: | 2011-08-23 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The molecular basis for the endocytosis of small R-SNAREs by the clathrin adaptor CALM.
Cell, 147, 2011
|
|
3ZYK
 
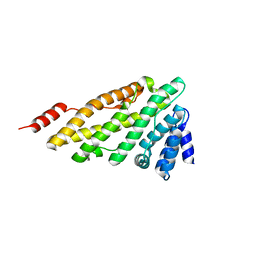 | | Structure of CALM (PICALM) ANTH domain | | Descriptor: | PHOSPHATIDYLINOSITOL-BINDING CLATHRIN ASSEMBLY PROTEIN | | Authors: | Miller, S.E, Sahlender, D.A, Graham, S.C, Honing, S, Robinson, M.S, Peden, A.A, Owen, D.J. | | Deposit date: | 2011-08-23 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The molecular basis for the endocytosis of small R-SNAREs by the clathrin adaptor CALM.
Cell, 147, 2011
|
|
3ZYM
 
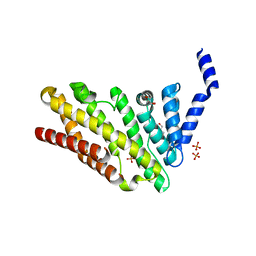 | | Structure of CALM (PICALM) in complex with VAMP8 | | Descriptor: | GLYCEROL, PHOSPHATE ION, PHOSPHATIDYLINOSITOL-BINDING CLATHRIN ASSEMBLY PROTEIN, ... | | Authors: | Miller, S.E, Sahlender, D.A, Graham, S.C, Honing, S, Robinson, M.S, Peden, A.A, Owen, D.J. | | Deposit date: | 2011-08-23 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The molecular basis for the endocytosis of small R-SNAREs by the clathrin adaptor CALM.
Cell, 147, 2011
|
|
6T5A
 
 | |
8A16
 
 | | Human PTPRM domains FN3-4, in spacegroup P212121 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Receptor-type tyrosine-protein phosphatase mu, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Caroe, E, Graham, S.C, Sharpe, H.J, Deane, J.E. | | Deposit date: | 2022-05-31 | | Release date: | 2023-03-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Determinants of receptor tyrosine phosphatase homophilic adhesion: Structural comparison of PTPRK and PTPRM extracellular domains.
J.Biol.Chem., 299, 2023
|
|
8A1F
 
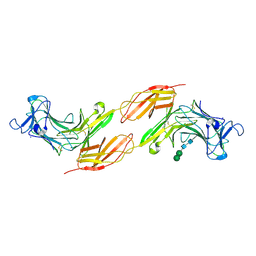 | | Human PTPRK N-terminal domains MAM-Ig-FN1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hay, I.M, Graham, S.C, Sharpe, H.J, Deane, J.E. | | Deposit date: | 2022-06-01 | | Release date: | 2023-03-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Determinants of receptor tyrosine phosphatase homophilic adhesion: Structural comparison of PTPRK and PTPRM extracellular domains.
J.Biol.Chem., 299, 2023
|
|
8A17
 
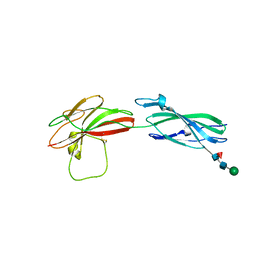 | | Human PTPRM domains FN3-4, in spacegroup P3221 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Receptor-type tyrosine-protein phosphatase mu, ... | | Authors: | Shamin, M, Graham, S.C, Sharpe, H.J, Deane, J.E. | | Deposit date: | 2022-05-31 | | Release date: | 2023-03-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Determinants of receptor tyrosine phosphatase homophilic adhesion: Structural comparison of PTPRK and PTPRM extracellular domains.
J.Biol.Chem., 299, 2023
|
|
6ZE9
 
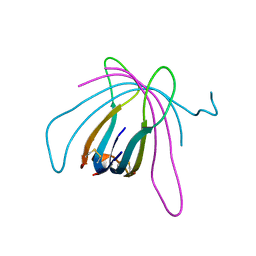 | |
7NBV
 
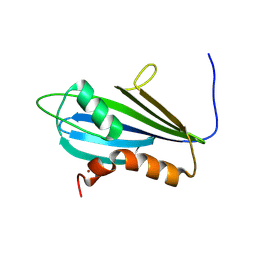 | | Structure of 2A protein from Theilers murine encephalomyelitis virus (TMEV) | | Descriptor: | BROMIDE ION, Capsid protein VP0 | | Authors: | Hill, C.H, Cook, G.M, Napthine, S, Kibe, A, Brown, K, Caliskan, N, Firth, A.E, Graham, S.C, Brierley, I. | | Deposit date: | 2021-01-28 | | Release date: | 2021-12-08 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Investigating molecular mechanisms of 2A-stimulated ribosomal pausing and frameshifting in Theilovirus.
Nucleic Acids Res., 49, 2021
|
|
7NWT
 
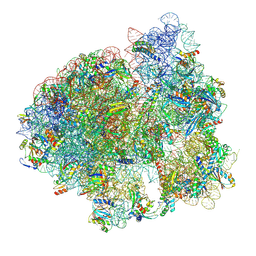 | | Initiated 70S ribosome in complex with 2A protein from encephalomyocarditis virus (EMCV) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Hill, C.H, Napthine, S, Pekarek, L, Kibe, A, Firth, A.E, Graham, S.C, Caliskan, N, Brierley, I. | | Deposit date: | 2021-03-17 | | Release date: | 2021-12-15 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structural and molecular basis for Cardiovirus 2A protein as a viral gene expression switch
Nat Commun, 12, 2021
|
|
5M5R
 
 | |
