1N2R
 
 | | A natural selected dimorphism in HLA B*44 alters self, peptide reportoire and T cell recognition. | | Descriptor: | ACETIC ACID, Beta-2-microglobulin, HLA DPA*0201 PEPTIDE, ... | | Authors: | Macdonald, W.A, Purcell, A.W, Williams, D.S, Mifsud, N, Ely, L.K, Gorman, J.J, Clements, C.S, Kjer-Nielsen, L, Koelle, D.M, Brooks, A.G, Lovrecz, G.O, Lu, L, Rossjohn, J, McCluskey, J. | | Deposit date: | 2002-10-24 | | Release date: | 2004-03-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A naturally selected dimorphism within the HLA-B44 supertype alters class I structure, peptide repertoire, and T cell recognition.
J.Exp.Med., 198, 2003
|
|
1XX9
 
 | | Crystal Structure of the FXIa Catalytic Domain in Complex with EcotinM84R | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Coagulation factor XI, Ecotin | | Authors: | Jin, L, Pandey, P, Babine, R.E, Gorga, J.C, Seidl, K.J, Gelfand, E, Weaver, D.T, Abdel-Meguid, S.S, Strickler, J.E. | | Deposit date: | 2004-11-04 | | Release date: | 2004-11-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of the FXIa Catalytic Domain in Complex with Ecotin Mutants Reveal Substrate-like Interactions
J.Biol.Chem., 280, 2005
|
|
1XXL
 
 | |
1XXD
 
 | | Crystal Structure of the FXIa Catalytic Domain in Complex with mutated Ecotin | | Descriptor: | Coagulation factor XI, Ecotin | | Authors: | Jin, L, Pandey, P, Babine, R.E, Gorga, J.C, Seidl, K.J, Gelfand, E, Weaver, D.T, Abdel-Meguid, S.S, Strickler, J.E. | | Deposit date: | 2004-11-04 | | Release date: | 2004-11-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Crystal Structures of the FXIa Catalytic Domain in Complex with Ecotin Mutants Reveal Substrate-like Interactions
J.Biol.Chem., 280, 2005
|
|
1XXF
 
 | | Crystal Structure of the FXIa Catalytic Domain in Complex with Ecotin Mutant (EcotinP) | | Descriptor: | Coagulation factor XI, Ecotin, SODIUM ION | | Authors: | Jin, L, Pandey, P, Babine, R.E, Gorga, J.C, Seidl, K.J, Gelfand, E, Weaver, D.T, Abdel-Meguid, S.S, Strickler, J.E. | | Deposit date: | 2004-11-04 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of the FXIa Catalytic Domain in Complex with Ecotin Mutants Reveal Substrate-like Interactions
J.Biol.Chem., 280, 2005
|
|
1Y9E
 
 | |
1Y7E
 
 | |
1P45
 
 | | Targeting tuberculosis and malaria through inhibition of enoyl reductase: compound activity and structural data | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN | | Authors: | Kuo, M.R, Morbidoni, H.R, Alland, D, Sneddon, S.F, Gourlie, B.B, Staveski, M.M, Leonard, M, Gregory, J.S, Janjigian, A.D, Yee, C, Musser, J.M, Kreiswirth, B.N, Iwamoto, H, Perozzo, R, Jacobs Jr, W.R, Sacchettini, J.C, Fidock, D.A, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-04-21 | | Release date: | 2003-09-16 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Targeting tuberculosis and malaria through inhibition of Enoyl reductase: compound activity and structural data.
J.Biol.Chem., 278, 2003
|
|
1P44
 
 | | Targeting tuberculosis and malaria through inhibition of enoyl reductase: compound activity and structural data | | Descriptor: | 5-{[4-(9H-FLUOREN-9-YL)PIPERAZIN-1-YL]CARBONYL}-1H-INDOLE, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Kuo, M.R, Morbidoni, H.R, Alland, D, Sneddon, S.F, Gourlie, B.B, Staveski, M.M, Leonard, M, Gregory, J.S, Janjigian, A.D, Yee, C, Musser, J.M, Kreiswirth, B, Iwamoto, H, Perozzo, R, Jacobs Jr, W.R, Sacchettini, J.C, Fidock, D.A, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-04-21 | | Release date: | 2003-09-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Targeting tuberculosis and malaria through inhibition of Enoyl reductase: compound activity and structural data.
J.Biol.Chem., 278, 2003
|
|
2YME
 
 | | Crystal structure of a mutant binding protein (5HTBP-AChBP) in complex with granisetron | | Descriptor: | 1-methyl-N-[(1R,5S)-9-methyl-9-azabicyclo[3.3.1]nonan-3-yl]indazole-3-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHATE ION, ... | | Authors: | Kesters, D, Thompson, A.J, Brams, M, Elk, R.v, Spurny, R, Geitmann, M, Villalgordo, J.M, Guskov, A, Danielson, U.H, Lummis, S.C.R, Smit, A.B, Ulens, C. | | Deposit date: | 2012-10-09 | | Release date: | 2012-12-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Ligand Recognition in 5-Ht3 Receptors.
Embo Rep., 14, 2013
|
|
2YMD
 
 | | Crystal structure of a mutant binding protein (5HTBP-AChBP) in complex with serotonin (5-hydroxytryptamine) | | Descriptor: | GLYCEROL, PHOSPHATE ION, SEROTONIN, ... | | Authors: | Kesters, D, Thompson, A.J, Brams, M, Elk, R.v, Spurny, R, Geitmann, M, Villalgordo, J.M, Guskov, A, Danielson, U.H, Lummis, S.C.R, Smit, A.B, Ulens, C. | | Deposit date: | 2012-10-09 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural Basis of Ligand Recognition in 5-Ht(3) Receptors.
Embo Rep., 14, 2013
|
|
4HPY
 
 | | Crystal structure of RV144-elicited antibody CH59 in complex with V2 peptide | | Descriptor: | CH59 Fab heavy chain, CH59 Fab light chain, Envelope glycoprotein gp160, ... | | Authors: | McLellan, J.S, Gorman, J, Haynes, B.F, Kwong, P.D. | | Deposit date: | 2012-10-24 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Vaccine Induction of Antibodies against a Structurally Heterogeneous Site of Immune Pressure within HIV-1 Envelope Protein Variable Regions 1 and 2.
Immunity, 38, 2013
|
|
1ZML
 
 | | Crystal Structure of the Catalytic Domain of Factor XI in complex with (R)-1-(4-(4-(hydroxymethyl)-1,3,2-dioxaborolan-2-yl)phenethyl)guanidine | | Descriptor: | (R)-1-(4-(4-(HYDROXYMETHYL)-1,3,2-DIOXABOROLAN-2-YL)PHENETHYL)GUANIDINE, BICARBONATE ION, Coagulation factor XI, ... | | Authors: | Lazarova, T.I, Jin, L, Rynkiewicz, M.J, Gorga, J.C, Bibbins, F, Meyers, H.V, Babine, R.E, Strickler, J.E. | | Deposit date: | 2005-05-10 | | Release date: | 2006-05-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Synthesis and in vitro biological evaluation of aryl boronic acids as potential inhibitors of factor XIa.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
1ZPB
 
 | | Crystal Structure of the Catalytic Domain of Coagulation Factor XI in Complex with 4-Methyl-pentanoic acid {1-[4-guanidino-1-(thiazole-2-carbonyl)-butylcarbamoyl]-2-methyl-propyl}-amide | | Descriptor: | 4-METHYL-PENTANOIC ACID {1-[4-GUANIDINO-1-(THIAZOLE-2-CARBONYL)-BUTYLCARBAMOYL]-2-METHYL-PROPYL}-AMIDE, Coagulation factor XI, SULFATE ION | | Authors: | Deng, H, Bannister, T.D, Jin, L, Nagafuji, P, Celatka, C.A, Lin, J, Lazarova, T.I, Rynkiewicz, M.J, Quinn, J, Bibbins, F, Pandey, P, Gorga, J, Babine, R.E, Meyers, H.V, Abdel-Meguid, S.S, Strickler, J.E. | | Deposit date: | 2005-05-16 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Synthesis, SAR exploration, and X-ray crystal structures of factor XIa inhibitors containing an alpha-ketothiazole arginine
Bioorg.Med.Chem.Lett., 16, 2006
|
|
1ZPC
 
 | | Crystal Structure of the Catalytic Domain of Coagulation Factor XI in Complex with 2-[2-(3-Chloro-phenyl)-2-hydroxy-acetylamino]-N-[4-guanidino-1-(thiazole-2-carbonyl)-butyl]-3-methyl-butyramide | | Descriptor: | 2-[2-(3-CHLORO-PHENYL)-2-HYDROXY-ACETYLAMINO]-N-[4-GUANIDINO-1-(THIAZOLE-2-CARBONYL)-BUTYL]-3-METHYL-BUTYRAMIDE, Coagulation factor XI, SULFATE ION | | Authors: | Deng, H, Bannister, T.D, Jin, L, Nagafuji, P, Celatka, C.A, Lin, J, Lazarova, T.I, Rynkiewicz, M.J, Quinn, J, Bibbins, F, Pandey, P, Gorga, J, Babine, R.E, Meyers, H.V, Abdel-Meguid, S.S, Strickler, J.E. | | Deposit date: | 2005-05-16 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Synthesis, SAR exploration, and X-ray crystal structures of factor XIa inhibitors containing an alpha-ketothiazole arginine
Bioorg.Med.Chem.Lett., 16, 2006
|
|
4HPO
 
 | | Crystal structure of RV144-elicited antibody CH58 in complex with V2 peptide | | Descriptor: | CH58 Fab heavy chain, CH58 Fab light chain, Envelope glycoprotein gp160, ... | | Authors: | McLellan, J.S, Gorman, J, Haynes, B.F, Kwong, P.D. | | Deposit date: | 2012-10-24 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.694 Å) | | Cite: | Vaccine Induction of Antibodies against a Structurally Heterogeneous Site of Immune Pressure within HIV-1 Envelope Protein Variable Regions 1 and 2.
Immunity, 38, 2013
|
|
1Q7E
 
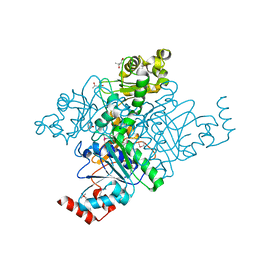 | | Crystal Structure of YfdW protein from E. coli | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Hypothetical protein yfdW, METHIONINE | | Authors: | Gogos, A, Gorman, J, Shapiro, L, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-08-18 | | Release date: | 2003-09-30 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Escherichia coli YfdW, a type III CoA transferase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1ZRK
 
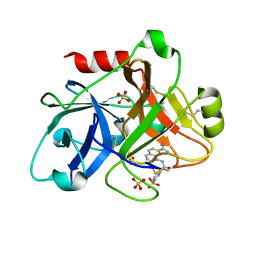 | | Factor XI complexed with 3-hydroxypropyl 3-(7-amidinonaphthalene-1-carboxamido)benzenesulfonate | | Descriptor: | 3-HYDROXYPROPYL 3-[({7-[AMINO(IMINO)METHYL]-1-NAPHTHYL}AMINO)CARBONYL]BENZENESULFONATE, Coagulation factor XI, SULFATE ION | | Authors: | Guo, Z, Bannister, T, Noll, R, Jin, L, Rynkiewicz, M.J, Bibbins, F, Magee, S, Gorga, J, Babine, R, Strickler, J. | | Deposit date: | 2005-05-19 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Synthesis and Optimization of Potent and Selective Inhibitors for Human Factor XIa: Substituted Naphthamidine Series
To be Published
|
|
1ZPZ
 
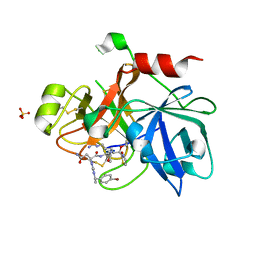 | | Factor XI catalytic domain complexed with N-((R)-1-(4-bromophenyl)ethyl)urea-Asn-Val-Arg-alpha-ketothiazole | | Descriptor: | Coagulation factor XI, N~2~-({[(1R)-1-(4-BROMOPHENYL)ETHYL]AMINO}CARBONYL)ASPARAGINYL-N~1~-{4-{[AMINO(IMINO)METHYL]AMINO}-1-[2,3-DIHYDRO-1,3-THIAZOL-2-YL(HYDROXY)METHYL]BUTYL}VALINAMIDE, SULFATE ION | | Authors: | Lin, J, Deng, H, Jin, L, Prandey, P, Rynkiewicz, M.J, Bibbins, F, Cantin, S, Quinn, J, Magee, S, Gorga, J. | | Deposit date: | 2005-05-18 | | Release date: | 2006-05-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design, Synthesis and Biological Evaluation of Peptidomimetic FXIa Inhibitors
To be Published
|
|
1HSA
 
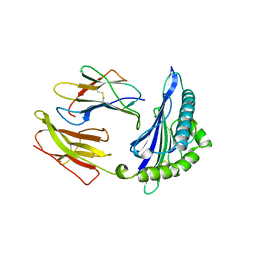 | | THE THREE-DIMENSIONAL STRUCTURE OF HLA-B27 AT 2.1 ANGSTROMS RESOLUTION SUGGESTS A GENERAL MECHANISM FOR TIGHT PEPTIDE BINDING TO MHC | | Descriptor: | BETA 2-MICROGLOBULIN, CLASS I HISTOCOMPATIBILITY ANTIGEN (HLA-B*2705), MODEL PEPTIDE SEQUENCE - ARAAAAAAA | | Authors: | Madden, D.R, Gorga, J.C, Strominger, J.L, Wiley, D.C. | | Deposit date: | 1992-08-11 | | Release date: | 1992-10-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The three-dimensional structure of HLA-B27 at 2.1 A resolution suggests a general mechanism for tight peptide binding to MHC.
Cell(Cambridge,Mass.), 70, 1992
|
|
1ZSJ
 
 | | Crystal Structure of the Catalytic Domain of Coagulation Factor XI in complex with N-(7-Carbamimidoyl-naphthalen-1-yl)-3-hydroxy-2-methyl-benzamide | | Descriptor: | BICARBONATE ION, Coagulation factor XI, N-(7-CARBAMIMIDOYL-NAPHTHALEN-1-YL)-3-HYDROXY-2-METHYL-BENZAMIDE, ... | | Authors: | Guo, Z, Bannister, T, Noll, R, Jin, L, Rynkiewicz, M, Bibbins, F, Magee, S, Gorga, J, Babine, R.E, Strickler, J.E, Meyers, H.V, Abdel-Meguid, S.S. | | Deposit date: | 2005-05-24 | | Release date: | 2006-05-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthesis and Optimization of Potent and Selective Inhibitors for Human Factor XIa: Substituted Naphthamidine Series
To be Published
|
|
1ZSK
 
 | | Crystal Structure of the Catalytic Domain of Coagulation Factor XI in Complex with 6-Carbamimidoyl-4-(3-hydroxy-2-methyl-benzoylamino)-naphthalene-2-carboxylic acid methyl ester | | Descriptor: | 6-CARBAMIMIDOYL-4-(3-HYDROXY-2-METHYL-BENZOYLAMINO)-NAPHTHALENE-2-CARBOXYLIC ACID METHYL ESTER, BICARBONATE ION, Coagulation factor XI, ... | | Authors: | Guo, Z, Bannister, T, Noll, R, Jin, L, Rynkiewicz, M, Bibbins, F, Magee, S, Gorga, J, Babine, R.E, Strickler, J.E, Meyers, H.V, Abdel-Meguid, S.S. | | Deposit date: | 2005-05-24 | | Release date: | 2006-05-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthesis and Optimization of Potent and Selective Inhibitors for Human Factor XIa: Substituted Naphthamidine Series
To be Published
|
|
1PQY
 
 | | Crystal structure of formyl-coA transferase yfdW from E. coli | | Descriptor: | Hypothetical protein yfdW | | Authors: | Gogos, A, Gorman, J, Shapiro, L, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-06-19 | | Release date: | 2003-09-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of Escherichia coli YfdW, a type III CoA transferase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1ICN
 
 | | ESCHERICHIA COLI-DERIVED RAT INTESTINAL FATTY ACID BINDING PROTEIN WITH BOUND MYRISTATE AT 1.5 A RESOLUTION AND I-FABPARG106-->GLN WITH BOUND OLEATE AT 1.74 A RESOLUTION | | Descriptor: | INTESTINAL FATTY ACID BINDING PROTEIN, OLEIC ACID | | Authors: | Eads, J.C, Sacchettini, J.C, Kromminga, A, Gordon, J.I. | | Deposit date: | 1993-09-20 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Escherichia coli-derived rat intestinal fatty acid binding protein with bound myristate at 1.5 A resolution and I-FABPArg106-->Gln with bound oleate at 1.74 A resolution.
J.Biol.Chem., 268, 1993
|
|
1ICM
 
 | | ESCHERICHIA COLI-DERIVED RAT INTESTINAL FATTY ACID BINDING PROTEIN WITH BOUND MYRISTATE AT 1.5 A RESOLUTION AND I-FABPARG106-->GLN WITH BOUND OLEATE AT 1.74 A RESOLUTION | | Descriptor: | INTESTINAL FATTY ACID BINDING PROTEIN, MYRISTIC ACID | | Authors: | Eads, J.C, Sacchettini, J.C, Kromminga, A, Gordon, J.I. | | Deposit date: | 1993-09-20 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Escherichia coli-derived rat intestinal fatty acid binding protein with bound myristate at 1.5 A resolution and I-FABPArg106-->Gln with bound oleate at 1.74 A resolution.
J.Biol.Chem., 268, 1993
|
|
