7KNH
 
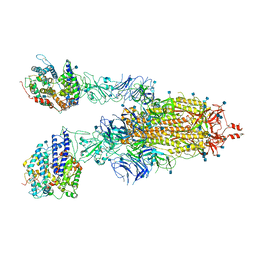 | | Cryo-EM Structure of Double ACE2-Bound SARS-CoV-2 Trimer Spike at pH 5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Gorman, J, Rapp, M, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-11-04 | | Release date: | 2020-12-16 | | Last modified: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
7KNB
 
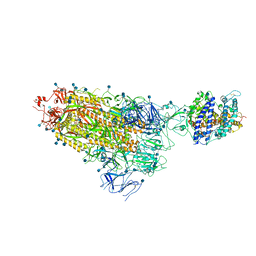 | | Cryo-EM structure of single ACE2-bound SARS-CoV-2 trimer spike at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Gorman, J, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-11-04 | | Release date: | 2020-12-09 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
7KJP
 
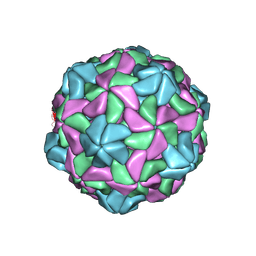 | | Disulfide Stabilized Norovirus GI.1 VLP Shell Region | | Descriptor: | Capsid protein VP1 | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2020-10-26 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Disulfide stabilization of human norovirus GI.1 virus-like particles focuses immune response toward blockade epitopes.
NPJ Vaccines, 5, 2020
|
|
7KMS
 
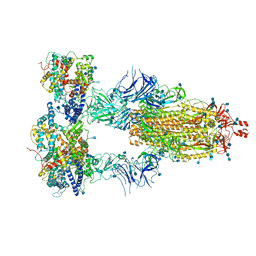 | | Cryo-EM structure of triple ACE2-bound SARS-CoV-2 trimer spike at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Gorman, J, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-11-03 | | Release date: | 2020-12-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
7KMZ
 
 | | Cryo-EM structure of double ACE2-bound SARS-CoV-2 trimer Spike at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Gorman, J, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-11-03 | | Release date: | 2020-12-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
6R2U
 
 | | Zinc-alpha2-Glycoprotein with a Fluorescent Dansyl C 11 Fatty Acid | | Descriptor: | 11-({[5-(dimethylamino)naphthalen-1-yl]sulfonyl}amino)undecanoic acid, AZIDE ION, SULFATE ION, ... | | Authors: | Lau, A.M, Gor, J, Perkins, S.J, Coker, A.R, McDermott, L.C. | | Deposit date: | 2019-03-18 | | Release date: | 2019-09-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal structure of zinc-alpha 2-glycoprotein in complex with a fatty acid reveals multiple different modes of protein-lipid binding.
Biochem.J., 476, 2019
|
|
5CKQ
 
 | | CUB1-EGF-CUB2 domains of rat MASP-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Mannan-binding lectin serine protease 1, ... | | Authors: | Nan, R, Furze, C.M, Wright, D.W, Gor, J, Wallis, R, Perkins, S.J. | | Deposit date: | 2015-07-15 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.704 Å) | | Cite: | Flexibility in Mannan-Binding Lectin-Associated Serine Proteases-1 and -2 Provides Insight on Lectin Pathway Activation.
Structure, 25, 2017
|
|
5CIS
 
 | | The CUB1-EGF-CUB2 domains of rat MBL-associated serine protease-2 (MASP-2) bound to Ca2+ | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Mannan-binding lectin serine peptidase 2 | | Authors: | Nan, R, Furze, C.M, Wright, D.W, Gor, J, Wallis, R, Perkins, S.J. | | Deposit date: | 2015-07-13 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Flexibility in Mannan-Binding Lectin-Associated Serine Proteases-1 and -2 Provides Insight on Lectin Pathway Activation.
Structure, 25, 2017
|
|
5CKM
 
 | | The CUB1-EGF-CUB2 domains of rat MBL-associated serine protease-2 (MASP-2) bound to Ca2+ | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Mannan-binding lectin serine peptidase 2 | | Authors: | Nan, R, Furze, C.M, Wright, D.W, Gor, J, Wallis, R, Perkins, S.J. | | Deposit date: | 2015-07-15 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Flexibility in Mannan-Binding Lectin-Associated Serine Proteases-1 and -2 Provides Insight on Lectin Pathway Activation.
Structure, 25, 2017
|
|
5CKN
 
 | | The CUB1-EGF-CUB2 domains of rat MBL-associated serine protease-2 (MASP-2) bound to Ca2+ | | Descriptor: | CALCIUM ION, Mannan-binding lectin serine peptidase 2 | | Authors: | Nan, R, Furze, C.M, Wright, D.W, Gor, J, Wallis, R, Perkins, S.J. | | Deposit date: | 2015-07-15 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Flexibility in Mannan-Binding Lectin-Associated Serine Proteases-1 and -2 Provides Insight on Lectin Pathway Activation.
Structure, 25, 2017
|
|
3GAU
 
 | |
3GAV
 
 | |
3GAW
 
 | |
3IRI
 
 | | Solution Structure of Heparin dp18 | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose | | Authors: | Khan, S, Gor, J, Mulloy, B, Perkins, S.J. | | Deposit date: | 2009-08-24 | | Release date: | 2009-11-03 | | Last modified: | 2024-02-21 | | Method: | SOLUTION SCATTERING | | Cite: | Semi-rigid solution structures of heparin by constrained X-ray scattering modelling: new insight into heparin-protein complexes.
J.Mol.Biol., 395, 2010
|
|
3IRL
 
 | | Solution Structure of Heparin dp36 | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose | | Authors: | Khan, S, Gor, J, Mulloy, B, Perkins, S.J. | | Deposit date: | 2009-08-24 | | Release date: | 2009-11-03 | | Last modified: | 2024-02-21 | | Method: | SOLUTION SCATTERING | | Cite: | Semi-rigid solution structures of heparin by constrained X-ray scattering modelling: new insight into heparin-protein complexes.
J.Mol.Biol., 395, 2010
|
|
3IRJ
 
 | | Solution Structure of Heparin dp24 | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose | | Authors: | Khan, S, Gor, J, Mulloy, B, Perkins, S.J. | | Deposit date: | 2009-08-24 | | Release date: | 2009-11-03 | | Last modified: | 2024-02-21 | | Method: | SOLUTION SCATTERING | | Cite: | Semi-rigid solution structures of heparin by constrained X-ray scattering modelling: new insight into heparin-protein complexes.
J.Mol.Biol., 395, 2010
|
|
3IRK
 
 | | Solution Structure of Heparin dp30 | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose | | Authors: | Khan, S, Gor, J, Mulloy, B, Perkins, S.J. | | Deposit date: | 2009-08-24 | | Release date: | 2009-11-03 | | Last modified: | 2024-02-21 | | Method: | SOLUTION SCATTERING | | Cite: | Semi-rigid solution structures of heparin by constrained X-ray scattering modelling: new insight into heparin-protein complexes.
J.Mol.Biol., 395, 2010
|
|
1U59
 
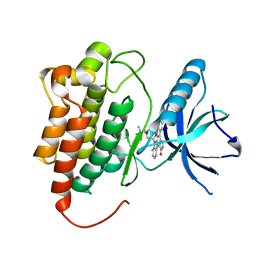 | | Crystal Structure of the ZAP-70 Kinase Domain in Complex with Staurosporine | | Descriptor: | STAUROSPORINE, Tyrosine-protein kinase ZAP-70 | | Authors: | Jin, L, Pluskey, S, Petrella, E.C, Cantin, S.M, Gorga, J.C, Rynkiewicz, M.J, Pandey, P, Strickler, J.E, Babine, R.E, Weaver, D.T, Seidl, K.J. | | Deposit date: | 2004-07-27 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Three-dimensional Structure of the ZAP-70 Kinase Domain in Complex with Staurosporine: IMPLICATIONS FOR THE DESIGN OF SELECTIVE INHIBITORS
J.Biol.Chem., 279, 2004
|
|
7S01
 
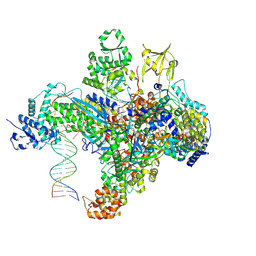 | | X-ray structure of the phage AR9 non-virion RNA polymerase holoenzyme in complex with a forked oligonucleotide containing the P077 promoter | | Descriptor: | DNA-directed RNA polymerase, DNA-directed RNA polymerase beta subunit, DNA-directed RNA polymerase beta' subunit, ... | | Authors: | Leiman, P.G, Sokolova, M.L, Gordeeva, J, Fraser, A, Severinov, K.V. | | Deposit date: | 2021-08-28 | | Release date: | 2022-07-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis of template strand deoxyuridine promoter recognition by a viral RNA polymerase.
Nat Commun, 13, 2022
|
|
1U6M
 
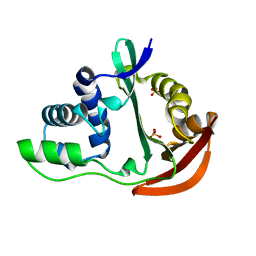 | | The crystal structure of acetyltransferase | | Descriptor: | SULFATE ION, acetyltransferase, GNAT family | | Authors: | Min, T, Gorman, J, Shapiro, L, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-07-30 | | Release date: | 2004-12-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of acetyltransferase, GNAT family from Enterococcus faecalis
To be Published
|
|
1TSJ
 
 | |
7RJF
 
 | | MOPD-1 mutant-L47W | | Descriptor: | MALONATE ION, ZINC ION, [L47W]MOPD-1 | | Authors: | Huawu, Y, Conan, K.W, Gordon, J.K, Brett, M.C, Yen-Hua, H, David, J.C. | | Deposit date: | 2021-07-20 | | Release date: | 2021-10-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rational Design of Potent Peptide Inhibitors of the PD-1:PD-L1 Interaction for Cancer Immunotherapy.
J.Am.Chem.Soc., 143, 2021
|
|
8VUE
 
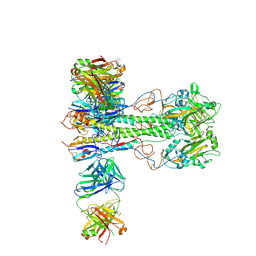 | | L5A7 Fab bound to Indonesia2005 Hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain, ... | | Authors: | Olia, A.S, Gorman, J, Kwong, P.D. | | Deposit date: | 2024-01-29 | | Release date: | 2024-05-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Anti-idiotype isolation of a broad and potent influenza A virus-neutralizing human antibody.
Front Immunol, 15, 2024
|
|
8G9W
 
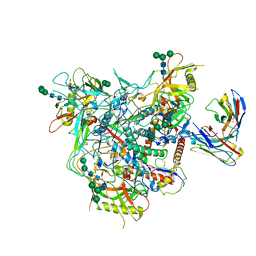 | | Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Changela, A, Gorman, J, Kwong, P.D. | | Deposit date: | 2023-02-22 | | Release date: | 2023-04-19 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (4.66 Å) | | Cite: | Diverse Murine Vaccinations Reveal Distinct Antibody Classes to Target Fusion Peptide and Variation in Peptide Length to Improve HIV Neutralization.
J.Virol., 97, 2023
|
|
8G9Y
 
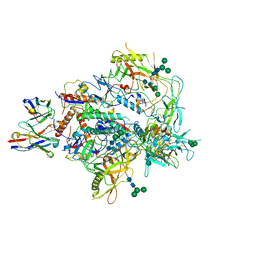 | | Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Changela, A, Gorman, J, Kwong, P.D. | | Deposit date: | 2023-02-22 | | Release date: | 2023-04-19 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (4.28 Å) | | Cite: | Diverse Murine Vaccinations Reveal Distinct Antibody Classes to Target Fusion Peptide and Variation in Peptide Length to Improve HIV Neutralization.
J.Virol., 97, 2023
|
|
