1YUP
 
 | | Reindeer beta-lactoglobulin | | Descriptor: | beta-lactoglobulin | | Authors: | Goldman, A, Oksanen, E. | | Deposit date: | 2005-02-14 | | Release date: | 2006-02-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Reindeer beta-lactoglobulin crystal structure with pseudo-body-centred noncrystallographic symmetry.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
3L2B
 
 | | Crystal structure of the CBS and DRTGG domains of the regulatory region of Clostridium perfringens pyrophosphatase complexed with activator, diadenosine tetraphosphate | | Descriptor: | BIS(ADENOSINE)-5'-TETRAPHOSPHATE, Probable manganase-dependent inorganic pyrophosphatase | | Authors: | Tuominen, H, Salminen, A, Oksanen, E, Jamsen, J, Heikkila, O, Lehtio, L, Magretova, N.N, Goldman, A, Baykov, A.A, Lahti, R. | | Deposit date: | 2009-12-15 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal Structures of the CBS and DRTGG Domains of the Regulatory Region of Clostridiumperfringens Pyrophosphatase Complexed with the Inhibitor, AMP, and Activator, Diadenosine Tetraphosphate.
J.Mol.Biol., 2010
|
|
1QHM
 
 | | ESCHERICHIA COLI PYRUVATE FORMATE LYASE LARGE DOMAIN | | Descriptor: | PYRUVATE FORMATE-LYASE | | Authors: | Leppanen, V.-M, Merckel, M.C, Ollis, D.L, Wong, K.K, Kozarich, J.W, Goldman, A. | | Deposit date: | 1999-05-19 | | Release date: | 2000-05-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Pyruvate formate lyase is structurally homologous to type I ribonucleotide reductase.
Structure Fold.Des., 7, 1999
|
|
8PRK
 
 | | THE R78K AND D117E ACTIVE SITE VARIANTS OF SACCHAROMYCES CEREVISIAE SOLUBLE INORGANIC PYROPHOSPHATASE: STRUCTURAL STUDIES AND MECHANISTIC IMPLICATIONS | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, PROTEIN (INORGANIC PYROPHOSPHATASE) | | Authors: | Tuominen, V, Heikinheimo, P, Kajander, T, Torkkel, T, Hyytia, T, Kapyla, J, Lahti, R, Cooperman, B.S, Goldman, A. | | Deposit date: | 1998-09-16 | | Release date: | 1998-12-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The R78K and D117E active-site variants of Saccharomyces cerevisiae soluble inorganic pyrophosphatase: structural studies and mechanistic implications.
J.Mol.Biol., 284, 1998
|
|
7O23
 
 | |
6ZH1
 
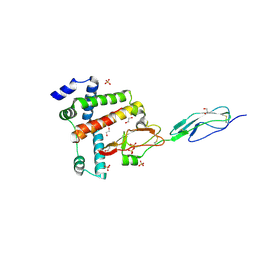 | | Crystal structure of complex between FH19-20 and FhbA protein from Borrelia hermsii | | Descriptor: | 1,2-ETHANEDIOL, Complement factor H, Factor H-binding protein A, ... | | Authors: | Kogan, K, Kotila, T, Meri, T, Goldman, A. | | Deposit date: | 2020-06-20 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of Borrelia immune evasion by FhbA-related proteins.
Plos Pathog., 18, 2022
|
|
2AWW
 
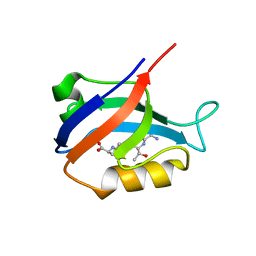 | | Synapse associated protein 97 PDZ2 domain variant C378G with C-terminal GluR-A peptide | | Descriptor: | 18-residue C-terminal peptide from glutamate receptor, ionotropic, AMPA1, ... | | Authors: | Von Ossowski, I, Oksanen, E, Von Ossowski, L, Cai, C, Sundberg, M, Goldman, A, Keinanen, K. | | Deposit date: | 2005-09-02 | | Release date: | 2006-08-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of the second PDZ domain of SAP97 in complex with a GluR-A C-terminal peptide
Febs J., 273, 2006
|
|
1IPW
 
 | |
5LNL
 
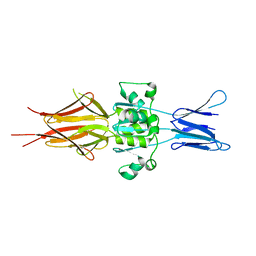 | | Crystal structure of Hsf 1608-1749 putative domain 1 | | Descriptor: | Hsf | | Authors: | Thomsen, M, Wright, J, Ridley, J, Goldman, A. | | Deposit date: | 2016-08-05 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The crystal structure of PD1, a Haemophilus surface fibril domain.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
3MUC
 
 | | MUCONATE CYCLOISOMERASE VARIANT I54V | | Descriptor: | MANGANESE (II) ION, PROTEIN (MUCONATE CYCLOISOMERASE) | | Authors: | Schell, U, Helin, S, Kajander, T, Schlomann, M, Goldman, A. | | Deposit date: | 1998-10-27 | | Release date: | 1999-11-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the activity of two muconate cycloisomerase variants toward substituted muconates.
Proteins, 34, 1999
|
|
6QP4
 
 | | Structure of 299-452 fragment of the UspA1 protein from Moraxella catarrhalis | | Descriptor: | CHLORIDE ION, HEXANE-1,6-DIOL, UspA1, ... | | Authors: | Mikula, K.M, Kolodziejczyk, R, Goldman, A. | | Deposit date: | 2019-02-13 | | Release date: | 2019-08-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the UspA1 protein fragment from Moraxella catarrhalis responsible for C3d binding.
J.Struct.Biol., 208, 2019
|
|
6QXA
 
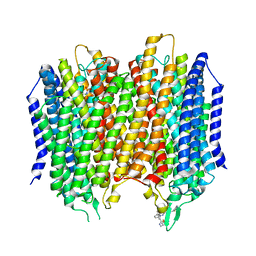 | | Structure of membrane bound pyrophosphatase from Thermotoga maritima in complex with imidodiphosphate and N-[(2-amino-6-benzothiazolyl)methyl]-1H-indole-2-carboxamide (ATC) | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, IMIDODIPHOSPHORIC ACID, K(+)-stimulated pyrophosphate-energized sodium pump, ... | | Authors: | Vidilaseris, K, Goldman, A. | | Deposit date: | 2019-03-07 | | Release date: | 2019-04-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Asymmetry in catalysis byThermotoga maritimamembrane-bound pyrophosphatase demonstrated by a nonphosphorus allosteric inhibitor.
Sci Adv, 5, 2019
|
|
2MUC
 
 | | MUCONATE CYCLOISOMERASE VARIANT F329I | | Descriptor: | MANGANESE (II) ION, PROTEIN (MUCONATE CYCLOISOMERASE) | | Authors: | Schell, U, Helin, S, Kajander, T, Schlomann, M, Goldman, A. | | Deposit date: | 1998-10-26 | | Release date: | 1999-12-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the activity of two muconate cycloisomerase variants toward substituted muconates.
Proteins, 34, 1999
|
|
4MUC
 
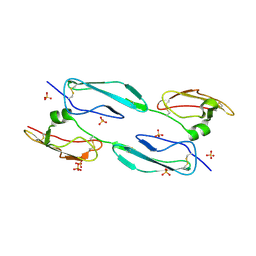 | | The 4th and 5th C-terminal domains of Factor H related protein 1 | | Descriptor: | Complement factor H-related protein 1, SULFATE ION | | Authors: | Bhattacharjee, A, Goldman, A, Kolodziejczyk, R, Jokiranta, T.S. | | Deposit date: | 2013-09-21 | | Release date: | 2015-02-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.897 Å) | | Cite: | The Major Autoantibody Epitope on Factor H in Atypical Hemolytic Uremic Syndrome Is Structurally Different from Its Homologous Site in Factor H-related Protein 1, Supporting a Novel Model for Induction of Autoimmunity in This Disease.
J.Biol.Chem., 290, 2015
|
|
1JOF
 
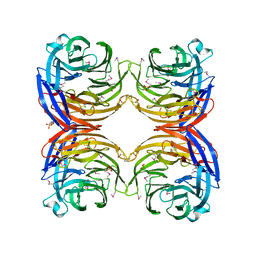 | | Neurospora crassa 3-carboxy-cis,cis-mucoante lactonizing enzyme | | Descriptor: | BETA-MERCAPTOETHANOL, CARBOXY-CIS,CIS-MUCONATE CYCLASE, PIPERAZINE-N,N'-BIS(2-ETHANESULFONIC ACID), ... | | Authors: | Kajander, T, Merckel, M.C, Thompson, A, Deacon, A.M, Mazur, P, Kozarich, J.W, Goldman, A. | | Deposit date: | 2001-07-28 | | Release date: | 2002-04-12 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of Neurospora crassa 3-carboxy-cis,cis-muconate lactonizing enzyme, a beta propeller cycloisomerase.
Structure, 10, 2002
|
|
8B22
 
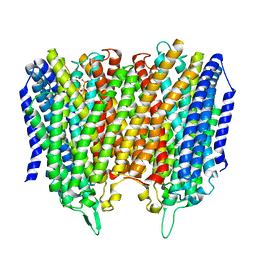 | |
8B23
 
 | |
8B21
 
 | | Time-resolved structure of K+-dependent Na+-PPase from Thermotoga maritima 0-60-seconds post reaction initiation with Na+ | | Descriptor: | DI(HYDROXYETHYL)ETHER, DODECYL-BETA-D-MALTOSIDE, K(+)-stimulated pyrophosphate-energized sodium pump, ... | | Authors: | Strauss, J, Vidilaseris, K, Goldman, A. | | Deposit date: | 2022-09-12 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Functional and structural asymmetry suggest a unifying principle for catalysis in membrane-bound pyrophosphatases.
Embo Rep., 25, 2024
|
|
8B37
 
 | | Crystal structure of Pyrobaculum aerophilum potassium-independent proton pumping membrane integral pyrophosphatase in complex with imidodiphosphate and magnesium, and with bound sulphate | | Descriptor: | IMIDODIPHOSPHORIC ACID, K(+)-insensitive pyrophosphate-energized proton pump, MAGNESIUM ION, ... | | Authors: | Strauss, J, Wilkinson, C, Vidilaseris, K, Ribeiro, O, Liu, J, Hillier, J, Malinen, A, Gehl, B, Jeuken, L.C, Pearson, A.R, Goldman, A. | | Deposit date: | 2022-09-16 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.84 Å) | | Cite: | Functional and structural asymmetry suggest a unifying principle for catalysis in membrane-bound pyrophosphatases.
Embo Rep., 25, 2024
|
|
8B24
 
 | | Time-resolved structure of K+-dependent Na+-PPase from Thermotoga maritima 3600-seconds post reaction initiation with Na+ | | Descriptor: | DIPHOSPHATE, K(+)-stimulated pyrophosphate-energized sodium pump, MAGNESIUM ION, ... | | Authors: | Strauss, J, Vidilaseris, K, Goldman, A. | | Deposit date: | 2022-09-12 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (4.53 Å) | | Cite: | Functional and structural asymmetry suggest a unifying principle for catalysis in membrane-bound pyrophosphatases.
Embo Rep., 25, 2024
|
|
2EIP
 
 | | INORGANIC PYROPHOSPHATASE | | Descriptor: | SOLUBLE INORGANIC PYROPHOSPHATASE | | Authors: | Kankare, J.A, Salminen, T, Goldman, A. | | Deposit date: | 1996-01-10 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Escherichia coli inorganic pyrophosphatase at 2.2 A resolution.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
3KZJ
 
 | | Structure of complement Factor H variant R1203A | | Descriptor: | Complement factor H, SULFATE ION | | Authors: | Bhattacharjee, A, Lehtinen, M.J, Kajander, T, Goldman, A, Jokiranta, T.S. | | Deposit date: | 2009-12-08 | | Release date: | 2010-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Both domain 19 and domain 20 of factor H are involved in binding to complement C3b and C3d
Mol.Immunol., 47, 2010
|
|
3KXV
 
 | | Structure of complement Factor H variant Q1139A | | Descriptor: | Complement factor H, SULFATE ION | | Authors: | Bhattacharjee, A, Lehtinen, M.J, Kajander, T, Goldman, A, Jokiranta, T.S. | | Deposit date: | 2009-12-04 | | Release date: | 2010-05-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Both domain 19 and domain 20 of factor H are involved in binding to complement C3b and C3d
Mol.Immunol., 47, 2010
|
|
3L31
 
 | | Crystal structure of the CBS and DRTGG domains of the regulatory region of Clostridium perfringens pyrophosphatase complexed with the inhibitor, AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Probable manganase-dependent inorganic pyrophosphatase | | Authors: | Tuominen, H, Salminen, A, Oksanen, E, Jamsen, J, Heikkila, O, Lehtio, L, Magretova, N.N, Goldman, A, Baykov, A.A, Lahti, R. | | Deposit date: | 2009-12-16 | | Release date: | 2010-04-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of the CBS and DRTGG Domains of the Regulatory Region of Clostridiumperfringens Pyrophosphatase Complexed with the Inhibitor, AMP, and Activator, Diadenosine Tetraphosphate.
J.Mol.Biol., 2010
|
|
2XQW
 
 | | Structure of Factor H domains 19-20 in complex with complement C3d | | Descriptor: | COMPLEMENT C3, COMPLEMENT FACTOR H | | Authors: | Kajander, T, Lehtinen, M.J, Hyvarinen, S, Bhattacharjee, A, Leung, E, Isenman, D.E, Meri, S, Jokiranta, T.S, Goldman, A. | | Deposit date: | 2010-09-07 | | Release date: | 2011-02-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.306 Å) | | Cite: | Dual Interaction of Factor H with C3D and Glycosaminoglycans in Host-Nonhost Discrimination by Complement.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
