6YUD
 
 | | Structure of Csx3/Crn3 from Archaeoglobus fulgidus in complex with cyclic tetra-adenylate (cA4) | | Descriptor: | Cyclic tetraadenosine monophosphate (cA4), Uncharacterized protein AF_1864 | | Authors: | McQuarrie, S, Gloster, T.M, White, M.F, Graham, S, Athukoralage, J.S, Gruschow, S. | | Deposit date: | 2020-04-27 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Tetramerisation of the CRISPR ring nuclease Crn3/Csx3 facilitates cyclic oligoadenylate cleavage.
Elife, 9, 2020
|
|
6SCE
 
 | | Structure of a Type III CRISPR defence DNA nuclease activated by cyclic oligoadenylate | | Descriptor: | Uncharacterized protein, cyclic oligoadenylate | | Authors: | McMahon, S.A, Zhu, W, Graham, S, White, M.F, Gloster, T.M. | | Deposit date: | 2019-07-24 | | Release date: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure and mechanism of a Type III CRISPR defence DNA nuclease activated by cyclic oligoadenylate.
Nat Commun, 11, 2020
|
|
6SCF
 
 | | A viral anti-CRISPR subverts type III CRISPR immunity by rapid degradation of cyclic oligoadenylate | | Descriptor: | Uncharacterized protein, cyclic oligoadenylate | | Authors: | McMahon, S.A, Athukoralage, J.S, Graham, S, White, M.F, Gloster, T.M. | | Deposit date: | 2019-07-24 | | Release date: | 2019-10-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | An anti-CRISPR viral ring nuclease subverts type III CRISPR immunity.
Nature, 577, 2020
|
|
4AD1
 
 | | Structure of the GH99 endo-alpha-mannosidase from Bacteroides xylanisolvens | | Descriptor: | GLYCOSYL HYDROLASE FAMILY 71 | | Authors: | Thompson, A.J, Williams, R.J, Hakki, Z, Alonzi, D.S, Wennekes, T, Gloster, T.M, Songsrirote, K, Thomas-Oates, J.E, Wrodnigg, T.M, Spreitz, J, Stuetz, A.E, Butters, T.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2011-12-21 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Mechanistic Insight Into N-Glycan Processing by Endo-Alpha-Mannosidase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4AD5
 
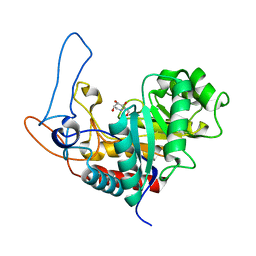 | | Structure of the GH99 endo-alpha-mannosidase from Bacteroides xylanisolvens in complex with glucose-1,3-deoxymannojirimycin and alpha-1,2-mannobiose | | Descriptor: | 1-DEOXYMANNOJIRIMYCIN, GLYCOSYL HYDROLASE FAMILY 71, alpha-D-glucopyranose, ... | | Authors: | Thompson, A.J, Williams, R.J, Hakki, Z, Alonzi, D.S, Wennekes, T, Gloster, T.M, Songsrirote, K, Thomas-Oates, J.E, Wrodnigg, T.M, Spreitz, J, Stuetz, A.E, Butters, T.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2011-12-21 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Mechanistic Insight Into N-Glycan Processing by Endo-Alpha-Mannosidase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4ACZ
 
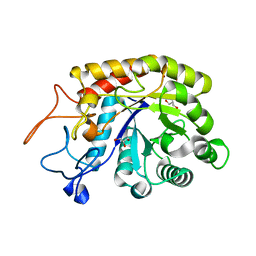 | | Structure of the GH99 endo-alpha-mannosidase from Bacteroides thetaiotaomicron | | Descriptor: | ENDO-ALPHA-MANNOSIDASE, GLYCEROL | | Authors: | Thompson, A.J, Williams, R.J, Hakki, Z, Alonzi, D.S, Wennekes, T, Gloster, T.M, Songsrirote, K, Thomas-Oates, J.E, Wrodnigg, T.M, Spreitz, J, Stuetz, A.E, Butters, T.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2011-12-21 | | Release date: | 2012-02-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural and Mechanistic Insight Into N-Glycan Processing by Endo-Alpha-Mannosidase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4AD2
 
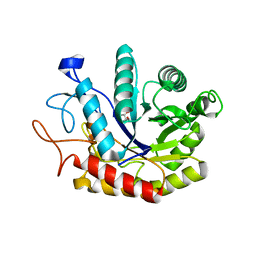 | | Structure of the GH99 endo-alpha-mannosidase from Bacteroides xylanisolvens in complex with glucose-1,3-isofagomine | | Descriptor: | 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, GLYCOSYL HYDROLASE FAMILY 71, alpha-D-glucopyranose | | Authors: | Thompson, A.J, Williams, R.J, Hakki, Z, Alonzi, D.S, Wennekes, T, Gloster, T.M, Songsrirote, K, Thomas-Oates, J.E, Wrodnigg, T.M, Spreitz, J, Stuetz, A.E, Butters, T.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2011-12-21 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Mechanistic Insight Into N-Glycan Processing by Endo-Alpha-Mannosidase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4AD3
 
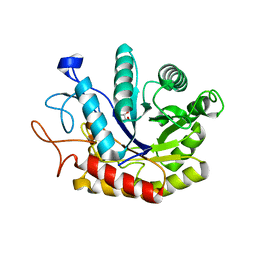 | | Structure of the GH99 endo-alpha-mannosidase from Bacteroides xylanisolvens in complex with Glucose-1,3-deoxymannojirimycin | | Descriptor: | 1-DEOXYMANNOJIRIMYCIN, GLYCOSYL HYDROLASE FAMILY 71, alpha-D-glucopyranose | | Authors: | Thompson, A.J, Williams, R.J, Hakki, Z, Alonzi, D.S, Wennekes, T, Gloster, T.M, Songsrirote, K, Thomas-Oates, J.E, Wrodnigg, T.M, Spreitz, J, Stuetz, A.E, Butters, T.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2011-12-21 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Mechanistic Insight Into N-Glycan Processing by Endo-Alpha-Mannosidase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4AD0
 
 | | Structure of the GH99 endo-alpha-mannosidase from Bacteriodes thetaiotaomicron in complex with BIS-TRIS-Propane | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ENDO-ALPHA-MANNOSIDASE, GLYCEROL | | Authors: | Thompson, A.J, Williams, R.J, Hakki, Z, Alonzi, D.S, Wennekes, T, Gloster, T.M, Songsrirote, K, Thomas-Oates, J.E, Wrodnigg, T.M, Spreitz, J, Stuetz, A.E, Butters, T.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2011-12-21 | | Release date: | 2012-02-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural and Mechanistic Insight Into N-Glycan Processing by Endo-Alpha-Mannosidase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4AD4
 
 | | Structure of the GH99 endo-alpha-mannosidase from Bacteroides xylanisolvens in complex with glucose-1,3-isofagomine and alpha-1,2- mannobiose | | Descriptor: | 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, GLYCOSYL HYDROLASE FAMILY 71, alpha-D-glucopyranose, ... | | Authors: | Thompson, A.J, Williams, R.J, Hakki, Z, Alonzi, D.S, Wennekes, T, Gloster, T.M, Songsrirote, K, Thomas-Oates, J.E, Wrodnigg, T.M, Spreitz, J, Stuetz, A.E, Butters, T.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2011-12-21 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Mechanistic Insight Into N-Glycan Processing by Endo-Alpha-Mannosidase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4ACY
 
 | | Selenomethionine derivative of the GH99 endo-alpha-mannosidase from Bacteroides thetaiotaomicron | | Descriptor: | ENDO-ALPHA-MANNOSIDASE, FORMIC ACID, GLYCEROL | | Authors: | Thompson, A.J, Williams, R.J, Hakki, Z, Alonzi, D.S, Wennekes, T, Gloster, T.M, Songsrirote, K, Thomas-Oates, J.E, Wrodnigg, T.M, Spreitz, J, Stuetz, A.E, Butters, T.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2011-12-21 | | Release date: | 2012-02-01 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural and Mechanistic Insight Into N-Glycan Processing by Endo-Alpha-Mannosidase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6ZZP
 
 | | Crystal structure of (R)-3-hydroxybutyrate dehydrogenase from Psychrobacter arcticus complexed with NAD+ and 3-oxovalerate | | Descriptor: | 3-oxidanylidenepentanoic acid, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative beta-hydroxybutyrate dehydrogenase | | Authors: | Machado, T.F.G, da Silva, R.G, Gloster, T.M, McMahon, S.A, Oehler, V. | | Deposit date: | 2020-08-04 | | Release date: | 2020-10-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Dissecting the Mechanism of ( R )-3-Hydroxybutyrate Dehydrogenase by Kinetic Isotope Effects, Protein Crystallography, and Computational Chemistry.
Acs Catalysis, 10, 2020
|
|
6ZZQ
 
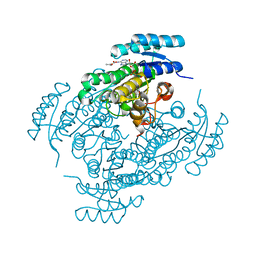 | | Crystal structure of (R)-3-hydroxybutyrate dehydrogenase from Acinetobacter baumannii complexed with NAD+ and acetoacetate | | Descriptor: | 3-hydroxybutyrate dehydrogenase, ACETOACETIC ACID, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Machado, T.F.G, da Silva, R.G, Gloster, T.M, McMahon, S.A, Oehler, V. | | Deposit date: | 2020-08-05 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Dissecting the Mechanism of ( R )-3-Hydroxybutyrate Dehydrogenase by Kinetic Isotope Effects, Protein Crystallography, and Computational Chemistry.
Acs Catalysis, 10, 2020
|
|
7BDV
 
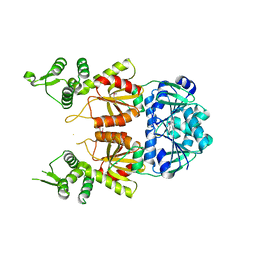 | | Structure of Can2 from Sulfobacillus thermosulfidooxidans in complex with cyclic tetra-adenylate (cA4) | | Descriptor: | Can2, Cyclic tetraadenosine monophosphate (cA4) | | Authors: | McQuarrie, S, McMahon, S.A, Gloster, T.M, White, M.F, Graham, S, Zhu, W, Gruschow, S. | | Deposit date: | 2020-12-22 | | Release date: | 2021-03-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The CRISPR ancillary effector Can2 is a dual-specificity nuclease potentiating type III CRISPR defence.
Nucleic Acids Res., 49, 2021
|
|
6ZZS
 
 | | Crystal structure of (R)-3-hydroxybutyrate dehydrogenase from Acinetobacter baumannii complexed with NAD+ and 3-oxovalerate | | Descriptor: | 3-hydroxybutyrate dehydrogenase, 3-oxidanylidenepentanoic acid, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Machado, T.F.G, da Silva, R.G, Gloster, T.M, McMahon, S.A, Oehler, V. | | Deposit date: | 2020-08-05 | | Release date: | 2020-10-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Dissecting the Mechanism of ( R )-3-Hydroxybutyrate Dehydrogenase by Kinetic Isotope Effects, Protein Crystallography, and Computational Chemistry.
Acs Catalysis, 10, 2020
|
|
6ZZO
 
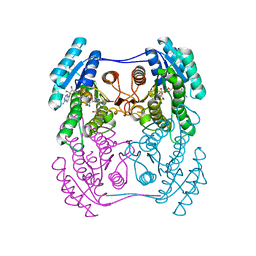 | | Crystal structure of (R)-3-hydroxybutyrate dehydrogenase from Psychrobacter arcticus complexed with NAD+ and acetoacetate | | Descriptor: | ACETOACETIC ACID, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative beta-hydroxybutyrate dehydrogenase | | Authors: | Machado, T.F.G, da Silva, R.G, Gloster, T.M, McMahon, S.A, Oehler, V. | | Deposit date: | 2020-08-04 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Dissecting the Mechanism of ( R )-3-Hydroxybutyrate Dehydrogenase by Kinetic Isotope Effects, Protein Crystallography, and Computational Chemistry.
Acs Catalysis, 10, 2020
|
|
4GZ3
 
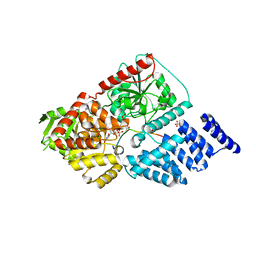 | | Crystal structure of human O-GlcNAc Transferase with UDP and a thioglycopeptide | | Descriptor: | 2-acetamido-2-deoxy-5-thio-beta-D-glucopyranose, Casein kinase II subunit alpha, SULFATE ION, ... | | Authors: | Lazarus, M.B, Jiang, J, Gloster, T.M, Zandberg, W.F, Vocadlo, D.J, Walker, S. | | Deposit date: | 2012-09-05 | | Release date: | 2012-10-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural snapshots of the reaction coordinate for O-GlcNAc transferase.
Nat.Chem.Biol., 8, 2012
|
|
4GZ6
 
 | | Crystal structure of human O-GlcNAc Transferase with UDP-5SGlcNAc | | Descriptor: | (2S,3R,4R,5S,6R)-3-(acetylamino)-4,5-dihydroxy-6-(hydroxymethyl)tetrahydro-2H-thiopyran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate, SULFATE ION, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit | | Authors: | Lazarus, M.B, Jiang, J, Gloster, T.M, Zandberg, W.F, Vocadlo, D.J, Walker, S. | | Deposit date: | 2012-09-06 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structural snapshots of the reaction coordinate for O-GlcNAc transferase.
Nat.Chem.Biol., 8, 2012
|
|
4GYY
 
 | | Crystal structure of human O-GlcNAc Transferase with UDP-5SGlcNAc and a peptide substrate | | Descriptor: | (2S,3R,4R,5S,6R)-3-(acetylamino)-4,5-dihydroxy-6-(hydroxymethyl)tetrahydro-2H-thiopyran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate, Casein kinase II subunit alpha, SULFATE ION, ... | | Authors: | Lazarus, M.B, Jiang, J, Gloster, T.M, Zandberg, W.F, Vocadlo, D.J, Walker, S. | | Deposit date: | 2012-09-05 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural snapshots of the reaction coordinate for O-GlcNAc transferase.
Nat.Chem.Biol., 8, 2012
|
|
2BM3
 
 | | Structure of the Type II cohesin from Clostridium thermocellum SdbA | | Descriptor: | ISOPROPYL ALCOHOL, SCAFFOLDING DOCKERIN BINDING PROTEIN A | | Authors: | Carvalho, A.L, Gloster, T.M, Pires, V.M.R, Proctor, M.R, Prates, J.A.M, Ferreira, L.M.A, Turkenburg, J.P, Romao, M.J, Davies, G.J, Gilbert, H.J, Fontes, C.M.G.A. | | Deposit date: | 2005-03-09 | | Release date: | 2005-03-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights Into the Structural Determinants of Cohesin-Dockerin Specificity Revealed by the Crystal Structure of the Type II Cohesin from Clostridium Thermocellum Sdba.
J.Mol.Biol., 349, 2005
|
|
2CC0
 
 | | Family 4 carbohydrate esterase from Streptomyces lividans in complex with acetate | | Descriptor: | ACETATE ION, ACETYL-XYLAN ESTERASE, ZINC ION | | Authors: | Taylor, E.J, Gloster, T.M, Turkenburg, J.P, Vincent, F, Brzozowski, A.M, Dupont, C, Shareck, F, Centeno, M.S.J, Prates, J.A.M, Puchart, V, Ferreira, L.M.A, Fontes, C.M.G.A, Biely, P, Davies, G.J. | | Deposit date: | 2006-01-10 | | Release date: | 2006-01-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and Activity of Two Metal-Ion Dependent Acetyl Xylan Esterases Involved in Plant Cell Wall Degradation Reveals a Close Similarity to Peptidoglycan Deacetylases
J.Biol.Chem., 281, 2006
|
|
2C79
 
 | | The structure of a family 4 acetyl xylan esterase from Clostridium thermocellum in complex with a colbalt ion. | | Descriptor: | COBALT (II) ION, GLYCOSIDE HYDROLASE, FAMILY 11:CLOSTRIDIUM CELLULOSOME ENZYME, ... | | Authors: | Taylor, E.J, Turkenburg, P.J, Vincent, F, Brzozowski, A.M, Gloster, T.M, Dupont, C, Shareck, F, Centeno, M.S.J, Prates, J.A.M, Ferreira, L.M.A, Fontes, C.M.G.A, Biely, P, Davies, G.J. | | Deposit date: | 2005-11-18 | | Release date: | 2006-01-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and Activity of Two Metal-Ion Dependent Acetyl Xylan Esterases Involved in Plant Cell Wall Degradation Reveals a Close Similarity to Peptidoglycan Deacetylases.
J.Biol.Chem., 281, 2006
|
|
2C71
 
 | | The structure of a family 4 acetyl xylan esterase from Clostridium thermocellum in complex with a magnesium ion. | | Descriptor: | GLYCOSIDE HYDROLASE, FAMILY 11:CLOSTRIDIUM CELLULOSOME ENZYME, DOCKERIN TYPE I:POLYSACCHARIDE, ... | | Authors: | Taylor, E.J, Turkenburg, P.J, Vincent, F, Brzozowski, A.M, Gloster, T.M, Dupont, C, Shareck, F, Centeno, M.S.J, Prates, J.A.M, Ferreira, L.M.A, Fontes, C.M.G.A, Biely, P, Davies, G.J. | | Deposit date: | 2005-11-17 | | Release date: | 2006-01-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structure and Activity of Two Metal-Ion Dependent Acetyl Xylan Esterases Involved in Plant Cell-Wall Degradation Reveals a Close Similarity to Peptidoglycan Deacetylases.
J.Biol.Chem., 281, 2006
|
|
2VRJ
 
 | | Beta-glucosidase from Thermotoga maritima in complex with N-octyl-5- deoxy-6-oxa-N-(thio)carbamoylcalystegine | | Descriptor: | (1S,2R,3S,4R,5R)-2,3,4-trihydroxy-N-octyl-6-oxa-8-azabicyclo[3.2.1]octane-8-carbothioamide, ACETATE ION, BETA-GLUCOSIDASE A, ... | | Authors: | Aguilar, M, Gloster, T.M, Garcia-Moreno, M.I, Ortiz Mellet, C, Davies, G.J, Llebaria, A, Casas, J, Egido-Gabas, M, Garcia Fernandez, J.M. | | Deposit date: | 2008-04-09 | | Release date: | 2008-10-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Basis for Beta-Glucosidase Inhibition by Ring-Modified Calystegine Analogues.
Chembiochem, 9, 2008
|
|
2WC4
 
 | | Structure of family 1 beta-glucosidase from Thermotoga maritima in complex with 3-imino-2-thia-(+)-castanospermine | | Descriptor: | (3Z,5S,6R,7S,8R,8aS)-3-(octylimino)hexahydro[1,3]thiazolo[3,4-a]pyridine-5,6,7,8-tetrol, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Aguilar, M, Gloster, T.M, Turkenburg, J.P, Garcia-Moreno, M.I, Ortiz Mellet, C, Davies, G.J, Garcia Fernandez, J.M. | | Deposit date: | 2009-03-08 | | Release date: | 2009-04-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Glycosidase Inhibition by Ring-Modified Castanospermine Analogues: Tackling Enzyme Selectivity by Inhibitor Tailoring.
Org.Biomol.Chem., 7, 2009
|
|
