6GWG
 
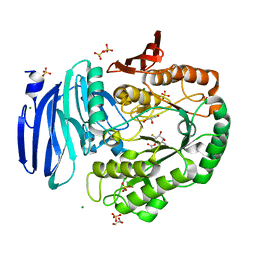 | | Alpha-galactosidase from Thermotoga maritima in complex with cyclohexene-based carbasugar mimic of galactose covalently linked to the nucleophile | | Descriptor: | (1~{S},2~{S},3~{S})-3-fluoranyl-6-(hydroxymethyl)cyclohex-5-ene-1,2,4-triol, Alpha-galactosidase, GLYCEROL, ... | | Authors: | Gloster, T.M, Oehler, V. | | Deposit date: | 2018-06-24 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Revealing the mechanism for covalent inhibition of glycoside hydrolases by carbasugars at an atomic level.
Nat Commun, 9, 2018
|
|
6GVD
 
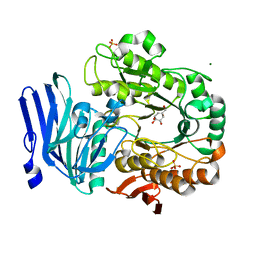 | | Alpha-galactosidase from Thermotoga maritima in complex with cyclohexene-based carbasugar mimic of galactose | | Descriptor: | (1~{S},2~{S},3~{S},4~{S})-5-(hydroxymethyl)cyclohex-5-ene-1,2,3,4-tetrol, Alpha-galactosidase, MAGNESIUM ION, ... | | Authors: | Gloster, T.M, Pengelly, R.J. | | Deposit date: | 2018-06-20 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Revealing the mechanism for covalent inhibition of glycoside hydrolases by carbasugars at an atomic level.
Nat Commun, 9, 2018
|
|
8C7F
 
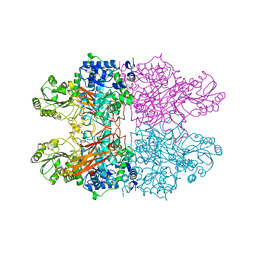 | |
7P1Q
 
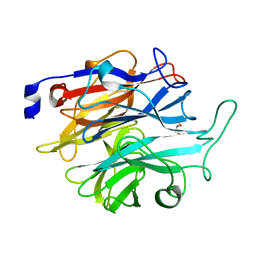 | |
7P1R
 
 | |
7P1D
 
 | |
7P1V
 
 | | Apo structure of KDNase from Trichophyton Rubrum | | Descriptor: | CALCIUM ION, Extracellular sialidase/neuraminidase, GLYCEROL | | Authors: | Gloster, T.M, McMahon, S.A. | | Deposit date: | 2021-07-02 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Kinetic and Structural Characterization of Sialidases (Kdnases) from Ascomycete Fungal Pathogens.
Acs Chem.Biol., 16, 2021
|
|
7ZB3
 
 | |
7ZEQ
 
 | |
7ZDY
 
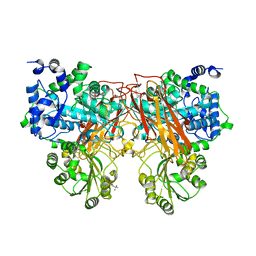 | |
7ZGZ
 
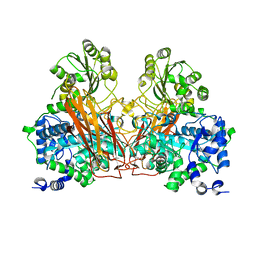 | |
6I6R
 
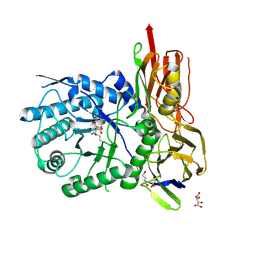 | | New Irreversible a-l-Iduronidase Inhibitors and Activity-Based Probes | | Descriptor: | (1~{R},2~{R},3~{R},4~{S},6~{S})-6-azanyl-2,3,4-tris(oxidanyl)cyclohexane-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-L-iduronidase, ... | | Authors: | Gloster, T.M, McMahon, S.A, Oehler, V. | | Deposit date: | 2018-11-15 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | New Irreversible alpha-l-Iduronidase Inhibitors and Activity-Based Probes.
Chemistry, 24, 2018
|
|
6I6X
 
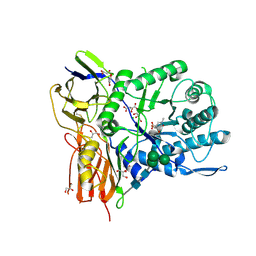 | | New Irreversible a-l-Iduronidase Inhibitors and Activity-Based Probes | | Descriptor: | (1~{R},2~{R},3~{R},4~{S},5~{S},6~{R})-7-methyl-3,4,5-tris(oxidanyl)-7-azabicyclo[4.1.0]heptane-2-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gloster, T.M, McMahon, S.A, Oehler, V. | | Deposit date: | 2018-11-15 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | New Irreversible alpha-l-Iduronidase Inhibitors and Activity-Based Probes.
Chemistry, 24, 2018
|
|
6GX8
 
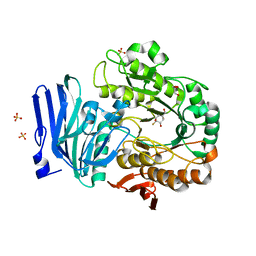 | | Alpha-galactosidase from Thermotoga maritima in complex with hydrolysed cyclohexene-based carbasugar mimic of galactose | | Descriptor: | (1~{S},2~{S},3~{S},4~{S})-3-fluoranyl-6-(hydroxymethyl)cyclohex-5-ene-1,2,4-triol, Alpha-galactosidase, GLYCEROL, ... | | Authors: | Gloster, T.M, Oehler, V. | | Deposit date: | 2018-06-26 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Revealing the mechanism for covalent inhibition of glycoside hydrolases by carbasugars at an atomic level.
Nat Commun, 9, 2018
|
|
6GTA
 
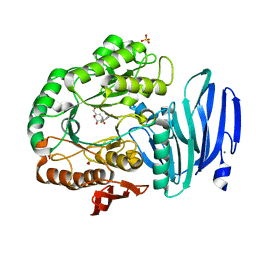 | | Alpha-galactosidase mutant D378A from Thermotoga maritima in complex with intact cyclohexene-based carbasugar mimic of galactose with 3,5 difluorophenyl leaving group | | Descriptor: | (1~{R},2~{S},3~{S},6~{S})-6-[3,5-bis(fluoranyl)phenoxy]-4-(hydroxymethyl)cyclohex-4-ene-1,2,3-triol, Alpha-galactosidase, MAGNESIUM ION, ... | | Authors: | Gloster, T.M, Pengelly, R.J. | | Deposit date: | 2018-06-17 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Revealing the mechanism for covalent inhibition of glycoside hydrolases by carbasugars at an atomic level.
Nat Commun, 9, 2018
|
|
6GWF
 
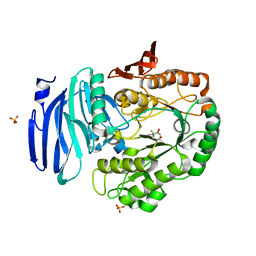 | | Alpha-galactosidase mutant D387A from Thermotoga maritima in complex with intact cyclohexene-based carbasugar mimic of galactose with 2,4-dinitro leaving group | | Descriptor: | (1~{S},2~{S},5~{S},6~{R})-5-(2,4-dinitrophenoxy)-6-fluoranyl-3-(hydroxymethyl)cyclohex-3-ene-1,2-diol, Alpha-galactosidase, MAGNESIUM ION, ... | | Authors: | Gloster, T.M, Oehler, V. | | Deposit date: | 2018-06-24 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Revealing the mechanism for covalent inhibition of glycoside hydrolases by carbasugars at an atomic level.
Nat Commun, 9, 2018
|
|
2CET
 
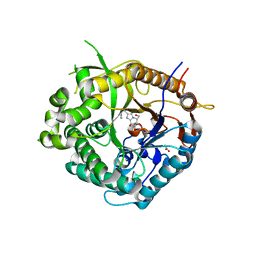 | | Beta-glucosidase from Thermotoga maritima in complex with phenethyl- substituted glucoimidazole | | Descriptor: | (5R,6R,7S,8S)-5-(HYDROXYMETHYL)-2-(2-PHENYLETHYL)-1,5,6,7,8,8A-HEXAHYDROIMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, ACETATE ION, BETA-GLUCOSIDASE A, ... | | Authors: | Gloster, T.M, Roberts, S, Vasella, A, Davies, G.J. | | Deposit date: | 2006-02-10 | | Release date: | 2006-09-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural, Kinetic, and Thermodynamic Analysis of Glucoimidazole-Derived Glycosidase Inhibitors.
Biochemistry, 45, 2006
|
|
2CBV
 
 | |
2CBU
 
 | | Beta-glucosidase from Thermotoga maritima in complex with castanospermine | | Descriptor: | ACETATE ION, BETA-GLUCOSIDASE A, CALCIUM ION, ... | | Authors: | Gloster, T.M, Davies, G.J, Madsen, R. | | Deposit date: | 2006-01-09 | | Release date: | 2006-02-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Dissection of Conformationally Restricted Inhibitors Binding to a Beta-Glucosidase.
Chembiochem, 7, 2006
|
|
2CES
 
 | | Beta-glucosidase from Thermotoga maritima in complex with glucoimidazole | | Descriptor: | ACETATE ION, BETA-GLUCOSIDASE A, CALCIUM ION, ... | | Authors: | Gloster, T.M, Roberts, S, Vasella, A, Davies, G.J. | | Deposit date: | 2006-02-10 | | Release date: | 2006-09-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural, Kinetic, and Thermodynamic Analysis of Glucoimidazole-Derived Glycosidase Inhibitors.
Biochemistry, 45, 2006
|
|
7O4O
 
 | | Structure of Staphylococcus aureus m1A22-tRNA methyltransferase in complex with S-adenosylhomocysteine | | Descriptor: | GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, tRNA (Adenine(22)-N(1))-methyltransferase | | Authors: | Gloster, T.M, Czekster, C.M, da Silva, R.G. | | Deposit date: | 2021-04-06 | | Release date: | 2022-04-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structure, dynamics, and molecular inhibition of the Staphylococcus aureus m 1 A22-tRNA methyltransferase TrmK.
J.Biol.Chem., 298, 2022
|
|
7O4N
 
 | | Structure of Staphylococcus aureus m1A22-tRNA methyltransferase in complex with S-adenosylmethionine | | Descriptor: | GLYCEROL, S-ADENOSYLMETHIONINE, tRNA (Adenine(22)-N(1))-methyltransferase | | Authors: | Gloster, T.M, Czekster, C.M, da Silva, R.G. | | Deposit date: | 2021-04-06 | | Release date: | 2022-04-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure, dynamics, and molecular inhibition of the Staphylococcus aureus m 1 A22-tRNA methyltransferase TrmK.
J.Biol.Chem., 298, 2022
|
|
7O4M
 
 | | Structure of Staphylococcus aureus m1A22-tRNA methyltransferase | | Descriptor: | CITRIC ACID, GLYCEROL, tRNA (Adenine(22)-N(1))-methyltransferase | | Authors: | Gloster, T.M, Czekster, C.M, da Silva, R.G. | | Deposit date: | 2021-04-06 | | Release date: | 2022-04-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure, dynamics, and molecular inhibition of the Staphylococcus aureus m 1 A22-tRNA methyltransferase TrmK.
J.Biol.Chem., 298, 2022
|
|
7P1E
 
 | | Structure of KDNase from Aspergillus Terrerus in complex with 2,3-difluoro-2-keto-3-deoxynononic acid | | Descriptor: | (2R,3R,4R,5R,6S)-2,3-bis(fluoranyl)-4,5-bis(oxidanyl)-6-[(1R,2R)-1,2,3-tris(oxidanyl)propyl]oxane-2-carboxylic acid, 3-deoxy-3-fluoro-D-erythro-alpha-L-manno-non-2-ulopyranosonic acid, CALCIUM ION, ... | | Authors: | Gloster, T.M, McMahon, S.A. | | Deposit date: | 2021-07-01 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Kinetic and Structural Characterization of Sialidases (Kdnases) from Ascomycete Fungal Pathogens.
Acs Chem.Biol., 16, 2021
|
|
7P1S
 
 | | Structure of KDNase from Trichophyton Rubrum in complex with 2,3-didehydro-2,3-dideoxy-D-glycero-D-galacto-nonulosonic acid. | | Descriptor: | 2,6-anhydro-3-deoxy-D-glycero-D-galacto-non-2-enonic acid, Extracellular sialidase/neuraminidase, SODIUM ION | | Authors: | Gloster, T.M, McMahon, S.A. | | Deposit date: | 2021-07-02 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Kinetic and Structural Characterization of Sialidases (Kdnases) from Ascomycete Fungal Pathogens.
Acs Chem.Biol., 16, 2021
|
|
