4HU7
 
 | | E. coli thioredoxin variant with Pro76 as single proline residue | | 分子名称: | COPPER (II) ION, SODIUM ION, Thioredoxin-1 | | 著者 | Glockshuber, R, Scharer, M.A, Capitani, G, Rubini, M. | | 登録日 | 2012-11-02 | | 公開日 | 2013-05-29 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | (4R)- and (4S)-Fluoroproline in the Conserved cis-Prolyl Peptide Bond of the Thioredoxin Fold: Tertiary Structure Context Dictates Ring Puckering.
Chembiochem, 14, 2013
|
|
6ZS5
 
 | | 3.5 A cryo-EM structure of human uromodulin filament core | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Uromodulin, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Stanisich, J.J, Zyla, D, Afanasyev, P, Xu, J, Pilhofer, M, Boeringer, D, Glockshuber, R. | | 登録日 | 2020-07-15 | | 公開日 | 2020-09-02 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | The cryo-EM structure of the human uromodulin filament core reveals a unique assembly mechanism.
Elife, 9, 2020
|
|
4W9Z
 
 | |
4WBR
 
 | |
4M8G
 
 | | Crystal structure of Se-Met hN33/Tusc3 | | 分子名称: | Tumor suppressor candidate 3 | | 著者 | Mohorko, E, Owen, R.L, Malojcic, G, Brozzo, M.S, Aebi, M, Glockshuber, R. | | 登録日 | 2013-08-13 | | 公開日 | 2014-03-26 | | 最終更新日 | 2014-05-07 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis of substrate specificity of human oligosaccharyl transferase subunit n33/tusc3 and its role in regulating protein N-glycosylation.
Structure, 22, 2014
|
|
4M90
 
 | | crystal structure of oxidized hN33/Tusc3 | | 分子名称: | Tumor suppressor candidate 3 | | 著者 | Mohorko, E, Owen, R.L, Malojcic, G, Brozzo, M.S, Aebi, M, Glockshuber, R. | | 登録日 | 2013-08-14 | | 公開日 | 2014-03-26 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural basis of substrate specificity of human oligosaccharyl transferase subunit n33/tusc3 and its role in regulating protein N-glycosylation.
Structure, 22, 2014
|
|
5OVW
 
 | | Nanobody-bound BtuF, the vitamin B12 binding protein in Escherichia coli | | 分子名称: | GLYCEROL, Nanobody, Vitamin B12-binding protein | | 著者 | Mireku, S.A, Sauer, M.M, Glockshuber, R, Locher, K.P. | | 登録日 | 2017-08-30 | | 公開日 | 2017-11-08 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.653 Å) | | 主引用文献 | Structural basis of nanobody-mediated blocking of BtuF, the cognate substrate-binding protein of the Escherichia coli vitamin B12 transporter BtuCD.
Sci Rep, 7, 2017
|
|
1JAE
 
 | | STRUCTURE OF TENEBRIO MOLITOR LARVAL ALPHA-AMYLASE | | 分子名称: | ALPHA-AMYLASE, CALCIUM ION, CHLORIDE ION | | 著者 | Strobl, S, Maskos, K, Betz, M, Wiegand, G, Huber, R, Gomis-Rueth, F.X, Frank, G, Glockshuber, R. | | 登録日 | 1997-09-30 | | 公開日 | 1998-11-04 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystal structure of yellow meal worm alpha-amylase at 1.64 A resolution.
J.Mol.Biol., 278, 1998
|
|
5LP9
 
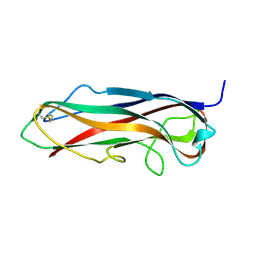 | | FimA wt from S. flexneri | | 分子名称: | Major type 1 subunit fimbrin (Pilin) | | 著者 | Zyla, D, Capitani, G, Prota, A, Glockshuber, R. | | 登録日 | 2016-08-12 | | 公開日 | 2017-12-20 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (0.88626635 Å) | | 主引用文献 | Alternative folding to a monomer or homopolymer is a common feature of the type 1 pilus subunit FimA from enteroinvasive bacteria.
J.Biol.Chem., 2019
|
|
2WCD
 
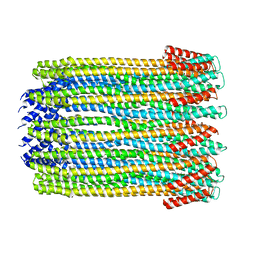 | | Crystal structure of the assembled cytolysin A pore | | 分子名称: | ETHYL MERCURY ION, HEMOLYSIN E, CHROMOSOMAL | | 著者 | Mueller, M, Grauschopf, U, Maier, T, Glockshuber, R, Ban, N. | | 登録日 | 2009-03-11 | | 公開日 | 2009-05-05 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.29 Å) | | 主引用文献 | The Structure of a Cytolytic Alpha-Helical Toxin Pore Reveals its Assembly Mechanism
Nature, 459, 2009
|
|
5NKT
 
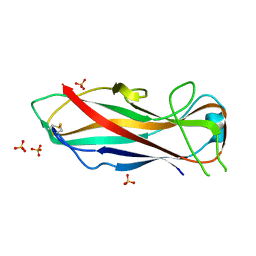 | | FimA wt from E. coli | | 分子名称: | SULFATE ION, Type-1 fimbrial protein, A chain | | 著者 | Zyla, D, Capitani, G, Prota, A, Glockshuber, R. | | 登録日 | 2017-04-03 | | 公開日 | 2018-05-16 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Alternative folding to a monomer or homopolymer is a common feature of the type 1 pilus subunit FimA from enteroinvasive bacteria.
J.Biol.Chem., 2019
|
|
3SQB
 
 | | Structure of the major type 1 pilus subunit FimA bound to the FimC chaperone | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Chaperone protein fimC, ... | | 著者 | Scharer, M.A, Eidam, O, Grutter, M.G, Glockshuber, R, Capitani, G. | | 登録日 | 2011-07-05 | | 公開日 | 2012-05-30 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Quality control of disulfide bond formation in pilus subunits by the chaperone FimC.
Nat.Chem.Biol., 8, 2012
|
|
1UN2
 
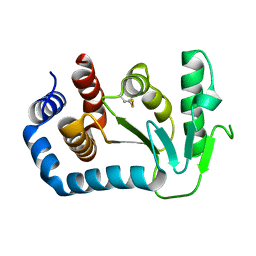 | | Crystal structure of circularly permuted CPDSBA_Q100T99: Preserved Global Fold and Local Structural Adjustments | | 分子名称: | THIOL-DISULFIDE INTERCHANGE PROTEIN | | 著者 | Manjasetty, B.A, Hennecke, J, Glockshuber, R, Heinemann, U. | | 登録日 | 2003-09-03 | | 公開日 | 2003-09-26 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structure of Circularly Permuted Dsba(Q100T99): Preserved Global Fold and Local Structural Adjustments
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1TMQ
 
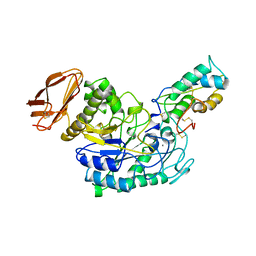 | | STRUCTURE OF TENEBRIO MOLITOR LARVAL ALPHA-AMYLASE IN COMPLEX WITH RAGI BIFUNCTIONAL INHIBITOR | | 分子名称: | CALCIUM ION, CHLORIDE ION, PROTEIN (ALPHA-AMYLASE), ... | | 著者 | Gomis-Rueth, F.X, Strobl, S, Glockshuber, R. | | 登録日 | 1998-01-13 | | 公開日 | 1999-03-02 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | A novel strategy for inhibition of alpha-amylases: yellow meal worm alpha-amylase in complex with the Ragi bifunctional inhibitor at 2.5 A resolution.
Structure, 6, 1998
|
|
7OT4
 
 | |
6Y7S
 
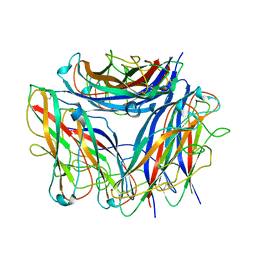 | | 2.85 A cryo-EM structure of the in vivo assembled type 1 pilus rod | | 分子名称: | Type-1 fimbrial protein, A chain | | 著者 | Zyla, D, Hospenthal, M, Waksman, G, Glockshuber, R. | | 登録日 | 2020-03-02 | | 公開日 | 2021-03-31 | | 最終更新日 | 2024-04-17 | | 実験手法 | ELECTRON MICROSCOPY (2.85 Å) | | 主引用文献 | The assembly platform FimD is required to obtain the most stable quaternary structure of type 1 pili.
Nat Commun, 15, 2024
|
|
4M92
 
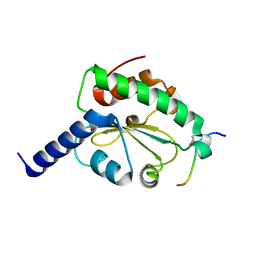 | | Crystal structure of hN33/Tusc3-peptide 2 | | 分子名称: | Interleukin-1 receptor accessory protein-like 1, Tumor suppressor candidate 3 | | 著者 | Mohorko, E, Owen, R.L, Malojcic, G, Brozzo, M.S, Aebi, M, Glockshuber, R. | | 登録日 | 2013-08-14 | | 公開日 | 2014-03-26 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural basis of substrate specificity of human oligosaccharyl transferase subunit n33/tusc3 and its role in regulating protein N-glycosylation.
Structure, 22, 2014
|
|
4M91
 
 | | crystal structure of hN33/Tusc3-peptide 1 | | 分子名称: | Protein cereblon, Tumor suppressor candidate 3 | | 著者 | Mohorko, E, Owen, R.L, Malojcic, G, Brozzo, M.S, Aebi, M, Glockshuber, R. | | 登録日 | 2013-08-14 | | 公開日 | 2014-03-26 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Structural basis of substrate specificity of human oligosaccharyl transferase subunit n33/tusc3 and its role in regulating protein N-glycosylation.
Structure, 22, 2014
|
|
4TXV
 
 | | Crystal structure of the mixed disulfide intermediate between thioredoxin-like TlpAs(C110S) and subunit II of cytochrome c oxidase CoxBPD (C233S) | | 分子名称: | Cytochrome c oxidase subunit 2, Thiol:disulfide interchange protein TlpA | | 著者 | Quade, N, Abicht, H.K, Hennecke, H, Glockshuber, R. | | 登録日 | 2014-07-07 | | 公開日 | 2014-10-01 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | How Periplasmic Thioredoxin TlpA Reduces Bacterial Copper Chaperone ScoI and Cytochrome Oxidase Subunit II (CoxB) Prior to Metallation.
J.Biol.Chem., 289, 2014
|
|
2MVX
 
 | | Atomic-resolution 3D structure of amyloid-beta fibrils: the Osaka mutation | | 分子名称: | Amyloid beta A4 protein | | 著者 | Schuetz, A.K, Vagt, T, Huber, M, Ovchinnikova, O.Y, Cadalbert, R, Wall, J, Guentert, P, Bockmann, A, Glockshuber, R, Meier, B.H. | | 登録日 | 2014-10-17 | | 公開日 | 2014-11-26 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLID-STATE NMR | | 主引用文献 | Atomic-Resolution Three-Dimensional Structure of Amyloid beta Fibrils Bearing the Osaka Mutation.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4TXO
 
 | | Crystal structure of the mixed disulfide complex of thioredoxin-like TlpAs(C110S) and copper chaperone ScoIs(C74S) | | 分子名称: | Blr1131 protein, DI(HYDROXYETHYL)ETHER, SODIUM ION, ... | | 著者 | Scharer, M.A, Abicht, H.K, Glockshuber, R, Hennecke, H. | | 登録日 | 2014-07-04 | | 公開日 | 2014-10-01 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | How Periplasmic Thioredoxin TlpA Reduces Bacterial Copper Chaperone ScoI and Cytochrome Oxidase Subunit II (CoxB) Prior to Metallation.
J.Biol.Chem., 289, 2014
|
|
3GA4
 
 | | Crystal structure of Ost6L (photoreduced form) | | 分子名称: | 1,2-ETHANEDIOL, Dolichyl-diphosphooligosaccharide-protein glycosyltransferase subunit OST6, TETRAETHYLENE GLYCOL | | 著者 | Stirnimann, C.U, Grimshaw, J.P.A, Schulz, B.L, Brozzo, M.S, Fritsch, F, Glockshuber, R, Capitani, G, Gruetter, M.G, Aebi, M. | | 登録日 | 2009-02-16 | | 公開日 | 2009-06-16 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Oxidoreductase activity of oligosaccharyltransferase subunits Ost3p and Ost6p defines site-specific glycosylation efficiency.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2M5G
 
 | | Solution structure of FimA wt | | 分子名称: | Type-1 fimbrial protein, A chain | | 著者 | Walczak, M.J, Puorger, C, Glockshuber, R, Wider, G. | | 登録日 | 2013-02-24 | | 公開日 | 2013-11-13 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Intramolecular donor strand complementation in the E. coli type 1 pilus subunit FimA explains the existence of FimA monomers as off-pathway products of pilus assembly that inhibit host cell apoptosis.
J.Mol.Biol., 426, 2014
|
|
3G9B
 
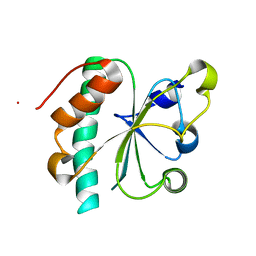 | | Crystal structure of reduced Ost6L | | 分子名称: | Dolichyl-diphosphooligosaccharide-protein glycosyltransferase subunit OST6 | | 著者 | Stirnimann, C.U, Grimshaw, J.P.A, Schulz, B.L, Brozzo, M.S, Fritsch, F, Glockshuber, R, Capitani, G, Gruetter, M.G, Aebi, M. | | 登録日 | 2009-02-13 | | 公開日 | 2009-06-16 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Oxidoreductase activity of oligosaccharyltransferase subunits Ost3p and Ost6p defines site-specific glycosylation efficiency.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1DSL
 
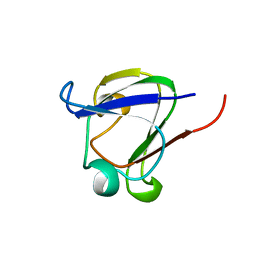 | | GAMMA B CRYSTALLIN C-TERMINAL DOMAIN | | 分子名称: | GAMMA B CRYSTALLIN | | 著者 | Norledge, B.V, Mayr, E.-M, Glockshuber, R, Bateman, O.A, Slingsby, C, Jaenicke, R, Driessen, H.P.C. | | 登録日 | 1996-02-01 | | 公開日 | 1996-07-11 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | The X-ray structures of two mutant crystallin domains shed light on the evolution of multi-domain proteins.
Nat.Struct.Biol., 3, 1996
|
|
