8SLV
 
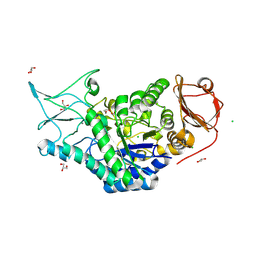 | | Structure of a salivary alpha-glucosidase from the mosquito vector Aedes aegypti. | | Descriptor: | 1,3-PROPANDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gittis, A.G, Williams, A.E, Garboczi, D, Calvo, E. | | Deposit date: | 2023-04-24 | | Release date: | 2024-03-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural and functional comparisons of salivary alpha-glucosidases from the mosquito vectors Aedes aegypti, Anopheles gambiae, and Culex quinquefasciatus.
Insect Biochem.Mol.Biol., 167, 2024
|
|
1NZN
 
 | |
5EJ0
 
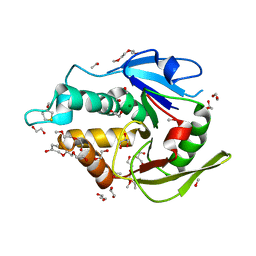 | | The vaccinia virus H3 envelope protein, a major target of neutralizing antibodies, exhibits a glycosyltransferase fold and binds UDP-Glucose | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, 2-(2-METHOXYETHOXY)ETHANOL, ... | | Authors: | Singh, K, Gittis, A.G, Gitti, R.K, Ostazesky, S.A, Su, H.P, Garboczi, D.N. | | Deposit date: | 2015-10-30 | | Release date: | 2016-03-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Vaccinia Virus H3 Envelope Protein, a Major Target of Neutralizing Antibodies, Exhibits a Glycosyltransferase Fold and Binds UDP-Glucose.
J.Virol., 90, 2016
|
|
6V4C
 
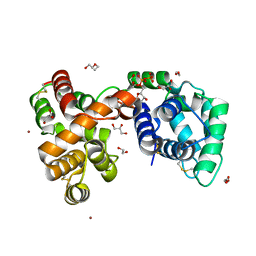 | | Culex quinquefasciatus D7 long form 1- CxD7L1 in complex with ADP | | Descriptor: | 1,2-ETHANEDIOL, 2-ETHOXYETHANOL, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Calvo, E, Garboczi, D.N, Martin-Martin, I, Gittis, A.G. | | Deposit date: | 2019-11-27 | | Release date: | 2020-06-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | ADP binding by the Culex quinquefasciatus mosquito D7 salivary protein enhances blood feeding on mammals.
Nat Commun, 11, 2020
|
|
7TXW
 
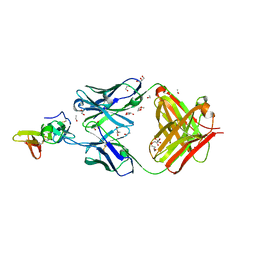 | | Crystal structure of the complex of the malaria sexual stage protein and vaccine target Pfs25 with the Fab fragment of a transmission blocking antibody 1G2 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ETHANOL, ... | | Authors: | Singh, K, Gittis, A.G, Garboczi, D.N. | | Deposit date: | 2022-02-10 | | Release date: | 2023-04-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural and immunological differences in Plasmodium falciparum sexual stage transmission-blocking vaccines comprised of Pfs25-EPA nanoparticles.
Npj Vaccines, 8, 2023
|
|
6OHG
 
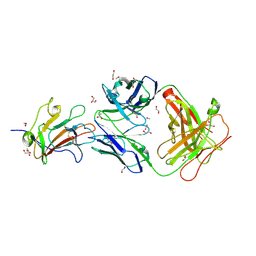 | | Structure of Plasmodium falciparum vaccine candidate Pfs230D1M in complex with the Fab of a transmission blocking antibody | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, 4F12 Heavy chain, ... | | Authors: | Garboczi, D.N, Singh, K, Gittis, A.G. | | Deposit date: | 2019-04-05 | | Release date: | 2020-06-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.385 Å) | | Cite: | Structure and function of a malaria transmission blocking vaccine targeting Pfs230 and Pfs230-Pfs48/45 proteins.
Commun Biol, 3, 2020
|
|
6CJ6
 
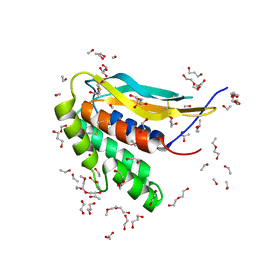 | | Structure of the poxvirus protein F9 | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, 2-(2-METHOXYETHOXY)ETHANOL, ... | | Authors: | Diesterbeck, U.S, Gittis, A.G, Garboczi, D.N, Moss, B. | | Deposit date: | 2018-02-26 | | Release date: | 2019-03-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The 2.1 angstrom structure of protein F9 and its comparison to L1, two components of the conserved poxvirus entry-fusion complex.
Sci Rep, 8, 2018
|
|
4QVB
 
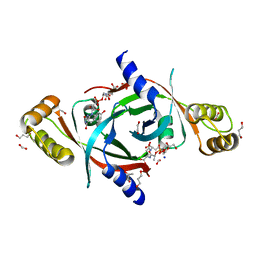 | | Mycobacterium tuberculosis protein Rv1155 in complex with co-enzyme F420 | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, COENZYME F420, ... | | Authors: | Mashalidis, E.H, Gittis, A.G, Tomczak, A, Abell, C, Barry III, C.E, Garboczi, D.N. | | Deposit date: | 2014-07-14 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular insights into the binding of coenzyme F420 to the conserved protein Rv1155 from Mycobacterium tuberculosis.
Protein Sci., 24, 2015
|
|
8U0R
 
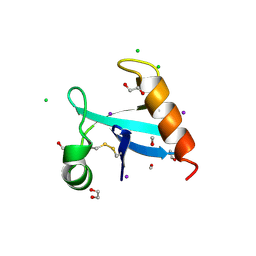 | | The crystal structure of protein A21, a component of the conserved poxvirus entry-fusion complex | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, 2-(2-METHOXYETHOXY)ETHANOL, ... | | Authors: | Diesterbeck, U, Gittis, A.G, Garboczi, D.N, Moss, B. | | Deposit date: | 2023-08-29 | | Release date: | 2024-09-04 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The 2.3 angstrom Structure of A21, a Protein Component of the Conserved Poxvirus Entry-Fusion Complex.
J.Mol.Biol., 437, 2025
|
|
7U1N
 
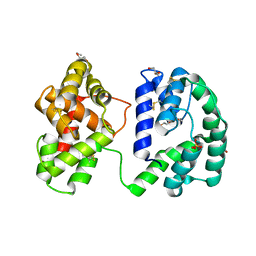 | | Crystal structure of the Anopheles darlingi AD-118 long form D7 salivary protein | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, 2-ETHOXYETHANOL, ... | | Authors: | Alvarenga, P.H, Gittis, A.G, Garboczi, D.N, Andersen, J.F. | | Deposit date: | 2022-02-21 | | Release date: | 2022-05-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Functional aspects of evolution in a cluster of salivary protein genes from mosquitoes.
Insect Biochem.Mol.Biol., 146, 2022
|
|
1HC8
 
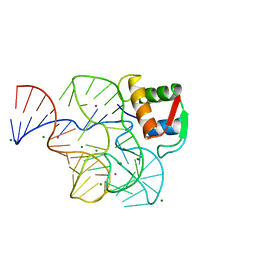 | | CRYSTAL STRUCTURE OF A CONSERVED RIBOSOMAL PROTEIN-RNA COMPLEX | | Descriptor: | 58 NUCLEOTIDE RIBOSOMAL 23S RNA DOMAIN, MAGNESIUM ION, OSMIUM ION, ... | | Authors: | Conn, G.L, Draper, D.E, Lattman, E.E, Gittis, A.G. | | Deposit date: | 2001-04-27 | | Release date: | 2002-05-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Compact RNA Tertiary Structure Contains a Buried Backbone-K+ Complex
J.Mol.Biol., 318, 2002
|
|
3BIS
 
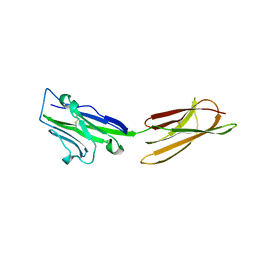 | | Crystal Structure of the PD-L1 | | Descriptor: | Programmed cell death 1 ligand 1 | | Authors: | Lin, D.Y, Tanaka, Y, Iwasaki, M, Gittis, A.G, Su, H.P, Mikami, B, Okazaki, T, Honjo, T, Minato, N, Garboczi, D.N. | | Deposit date: | 2007-11-30 | | Release date: | 2008-02-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | The PD-1/PD-L1 complex resembles the antigen-binding Fv domains of antibodies and T cell receptors.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3BIK
 
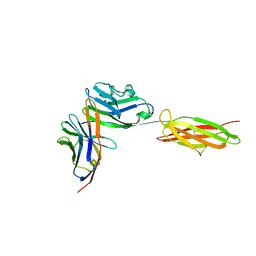 | | Crystal Structure of the PD-1/PD-L1 Complex | | Descriptor: | GLYCEROL, Programmed cell death 1 ligand 1, Programmed cell death protein 1 | | Authors: | Lin, D.Y, Tanaka, Y, Iwasaki, M, Gittis, A.G, Su, H.P, Mikami, B, Okazaki, T, Honjo, T, Minato, N, Garboczi, D.N. | | Deposit date: | 2007-11-30 | | Release date: | 2008-02-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The PD-1/PD-L1 complex resembles the antigen-binding Fv domains of antibodies and T cell receptors.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3C64
 
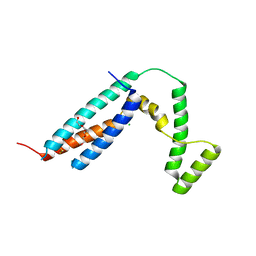 | | The MC179 portion of the Cysteine-rich Interdomain Region (CIDR) of a Plasmodium falciparum Erythrocyte Membrane Protein-1 (PfEMP1) | | Descriptor: | CHLORIDE ION, PfEMP1 variant 2 of strain MC, TETRAETHYLENE GLYCOL | | Authors: | Klein, M.M, Gittis, A.G, Su, H.P, Makobongo, M.O, Moore, J.M, Singh, S, Miller, L.H, Garboczi, D.N. | | Deposit date: | 2008-02-02 | | Release date: | 2008-09-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Cysteine-Rich Interdomain Region from the Highly Variable Plasmodium falciparum Erythrocyte Membrane Protein-1 Exhibits a Conserved Structure.
Plos Pathog., 4, 2008
|
|
2I9L
 
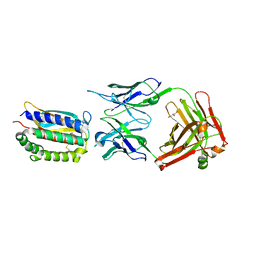 | | Structure of Fab 7D11 from a neutralizing antibody against the poxvirus L1 protein | | Descriptor: | Antibody 7D11 heavy chain, Antibody 7D11 light chain, GLYCEROL, ... | | Authors: | Su, H.P, Golden, J.W, Gittis, A.G, Moss, B, Hooper, J.W, Garboczi, D.N. | | Deposit date: | 2006-09-05 | | Release date: | 2007-09-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for the binding of the neutralizing antibody, 7D11, to the poxvirus L1 protein
Virology, 368, 2007
|
|
1ZJE
 
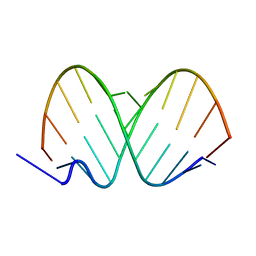 | | 12mer-spd | | Descriptor: | 5'-D(*AP*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*T)-3', 5'-D(*TP*AP*GP*CP*CP*CP*CP*GP*CP*CP*CP*C)-3' | | Authors: | Dohm, J.A, Hsu, M.H, Hwu, J.R, Huang, R.C, Moudrianakis, E.N, Lattman, E.E, Gittis, A.G. | | Deposit date: | 2005-04-28 | | Release date: | 2005-05-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Influence of Ions, Hydration, and the Transcriptional Inhibitor P4N on the Conformations of the Sp1 Binding Site.
J.Mol.Biol., 349, 2005
|
|
1ZJG
 
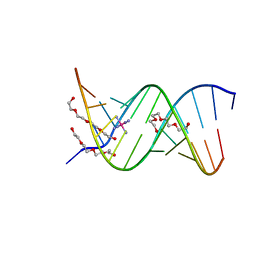 | | 13mer-co | | Descriptor: | 5'-D(*AP*TP*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*T)-3', 5'-D(*TP*AP*GP*CP*CP*CP*CP*GP*CP*CP*CP*CP*A)-3', COBALT HEXAMMINE(III), ... | | Authors: | Dohm, J.A, Hsu, M.H, Hwu, J.R, Huang, R.C, Moudrianakis, E.N, Lattman, E.E, Gittis, A.G. | | Deposit date: | 2005-04-28 | | Release date: | 2005-05-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Influence of Ions, Hydration, and the Transcriptional Inhibitor P4N on the Conformations of the Sp1 Binding Site.
J.Mol.Biol., 349, 2005
|
|
1ZJF
 
 | | 12mer-spd-P4N | | Descriptor: | 5'-D(*AP*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*T)-3', 5'-D(*TP*AP*GP*CP*CP*CP*CP*GP*CP*CP*CP*C)-3' | | Authors: | Dohm, J.A, Hsu, M.H, Hwu, J.R, Huang, R.C, Moudrianakis, E.N, Lattman, E.E, Gittis, A.G. | | Deposit date: | 2005-04-28 | | Release date: | 2005-05-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Influence of Ions, Hydration, and the Transcriptional Inhibitor P4N on the Conformations of the Sp1 Binding Site.
J.Mol.Biol., 349, 2005
|
|
1EX3
 
 | |
1GEN
 
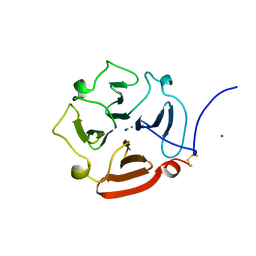 | | C-TERMINAL DOMAIN OF GELATINASE A | | Descriptor: | CALCIUM ION, CHLORIDE ION, GELATINASE A, ... | | Authors: | Libson, A.M, Gittis, A.G, Collier, I.E, Marmer, B.L, Goldberg, G.G, Lattman, E.E. | | Deposit date: | 1995-07-19 | | Release date: | 1996-08-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of the haemopexin-like C-terminal domain of gelatinase A.
Nat.Struct.Biol., 2, 1995
|
|
2OEO
 
 | |
3CML
 
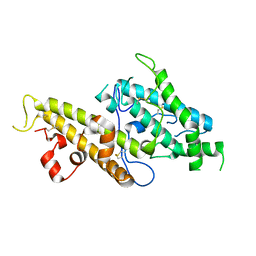 | | Crystal Structure of the DBL3x domain of the Plasmodium falcipurum VAR2CSA protein | | Descriptor: | Erythrocyte membrane protein 1 | | Authors: | Singh, K, Gittis, A.G, Nguyen, P, Gowda, D.C, Miller, L.H, Garboczi, D.N. | | Deposit date: | 2008-03-23 | | Release date: | 2008-09-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the DBL3x domain of pregnancy-associated malaria protein VAR2CSA complexed with chondroitin sulfate A.
Nat.Struct.Mol.Biol., 15, 2008
|
|
3CPZ
 
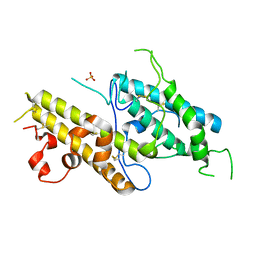 | | Crystal structure of VAR2CSA DBL3x domain in the presence of dodecasaccharide of CSA | | Descriptor: | Erythrocyte membrane protein 1, SULFATE ION | | Authors: | Singh, K, Gittis, A.G, Nguyen, P, Gowda, D.C, Miller, L.H, Garboczi, D.N. | | Deposit date: | 2008-04-01 | | Release date: | 2008-09-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the DBL3x domain of pregnancy-associated malaria protein VAR2CSA complexed with chondroitin sulfate A.
Nat.Struct.Mol.Biol., 15, 2008
|
|
2SOB
 
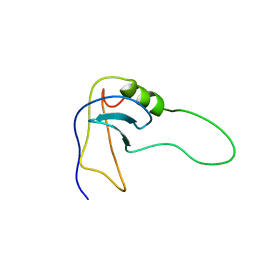 | | SN-OB, OB-FOLD SUB-DOMAIN OF STAPHYLOCOCCAL NUCLEASE, NMR, 10 STRUCTURES | | Descriptor: | STAPHYLOCOCCAL NUCLEASE | | Authors: | Alexandrescu, A.T, Gittis, A.G, Abeygunawardana, C, Shortle, D. | | Deposit date: | 1995-09-15 | | Release date: | 1995-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a stable "OB-fold" sub-domain isolated from staphylococcal nuclease.
J.Mol.Biol., 250, 1995
|
|
7KC8
 
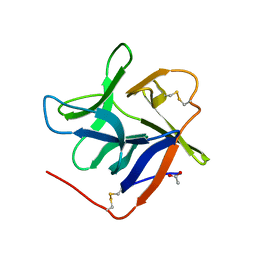 | | Salivary protein from Culex quinquefasciatus that belongs to the Cysteins and Tryptophan-Rich (CWRC) family | | Descriptor: | 16.4 kDa salivary peptide, ACETIC ACID | | Authors: | Garboczi, D.N, Gittis, A.G, Kern, O, Martin-Martin, I. | | Deposit date: | 2020-10-05 | | Release date: | 2021-06-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structures of two salivary proteins from the West Nile vector Culex quinquefasciatus reveal a beta-trefoil fold with putative sugar binding properties
Curr Res Struct Biol, 3, 2021
|
|
