8K6Y
 
 | | Serial femtosecond crystallography structure of photo dissociated CO from ba3- type cytochrome c oxidase determined by extrapolation method | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CARBON MONOXIDE, COPPER (II) ION, ... | | Authors: | Safari, C, Ghosh, S, Andersson, R, Johannesson, J, Donoso, A.V, Zoric, D, Sandelin, E, Iwata, S, Neutze, R, Branden, G. | | Deposit date: | 2023-07-25 | | Release date: | 2023-11-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Time-resolved serial crystallography to track the dynamics of carbon monoxide in the active site of cytochrome c oxidase.
Sci Adv, 9, 2023
|
|
8K65
 
 | | Serial femtosecond crystallography structure of CO bound ba3- type cytochrome c oxidase without pump laser irradiation | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CARBON MONOXIDE, COPPER (II) ION, ... | | Authors: | Safari, C, Ghosh, S, Andersson, R, Johannesson, J, Donoso, A.V, Bath, P, Zoric, D, Sandelin, E, Nango, E, Tanaka, R, Iwata, S, Neutze, R, Branden, G. | | Deposit date: | 2023-07-25 | | Release date: | 2023-11-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Time-resolved serial crystallography to track the dynamics of carbon monoxide in the active site of cytochrome c oxidase.
Sci Adv, 9, 2023
|
|
1RAM
 
 | | A NOVEL DNA RECOGNITION MODE BY NF-KB P65 HOMODIMER | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, DNA (5'-D(*CP*GP*GP*CP*TP*GP*GP*AP*AP*AP*TP*TP*TP*CP*CP*AP*GP*CP*CP*G)-3'), PROTEIN (TRANSCRIPTION FACTOR NF-KB P65) | | Authors: | Chen, Y.-Q, Ghosh, S, Ghosh, G. | | Deposit date: | 1997-11-22 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A novel DNA recognition mode by the NF-kappa B p65 homodimer.
Nat.Struct.Biol., 5, 1998
|
|
1DF1
 
 | | MURINE INOSOXY DIMER WITH ISOTHIOUREA BOUND IN THE ACTIVE SITE | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ETHYLISOTHIOUREA, NITRIC OXIDE SYNTHASE, ... | | Authors: | Crane, B.R, Rosenfeld, R.J, Arvai, A.S, Ghosh, D.K, Ghosh, S, Tainer, J.A, Stuehr, D.J, Getzoff, E.D. | | Deposit date: | 1999-11-16 | | Release date: | 1999-12-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | N-terminal domain swapping and metal ion binding in nitric oxide synthase dimerization.
EMBO J., 18, 1999
|
|
4PT4
 
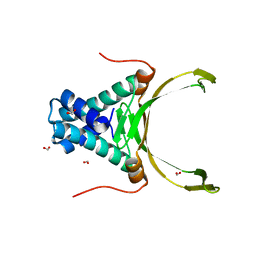 | | Crystal structure Analysis of N terminal region containing the dimerization domain and DNA binding domain of HU protein(Histone like protein-DNA binding) from Mycobacterium tuberculosis [H37Ra] | | Descriptor: | DNA-binding protein HU homolog, FORMIC ACID | | Authors: | Bhowmick, T, Ramagopal, U.A, Ghosh, S, Nagaraja, V, Ramakumar, S. | | Deposit date: | 2014-03-10 | | Release date: | 2014-05-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Targeting Mycobacterium tuberculosis nucleoid-associated protein HU with structure-based inhibitors
Nat Commun, 5, 2014
|
|
2XHB
 
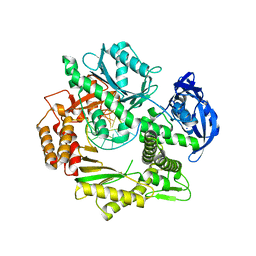 | | Crystal structure of DNA polymerase from Thermococcus gorgonarius in complex with hypoxanthine-containing DNA | | Descriptor: | 5'-D(*AP*AP*AP*AP*AP*AP*AP*AP*A)-3', DNA POLYMERASE, HYPOXANTHINE-CONTAINING DNA, ... | | Authors: | Killelea, T, Ghosh, S, Tan, S.S, Heslop, P, Firbank, S.J, Kool, E.T, Connolly, B.A. | | Deposit date: | 2010-06-14 | | Release date: | 2010-07-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Probing the Interaction of Archaeal DNA Polymerases with Deaminated Bases Using X-Ray Crystallography and Non-Hydrogen Bonding Isosteric Base Analogues.
Biochemistry, 49, 2010
|
|
6VDE
 
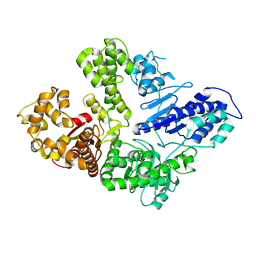 | | Full-length M. smegmatis Pol1 | | Descriptor: | DNA polymerase I, MANGANESE (II) ION | | Authors: | Shuman, S, Goldgur, Y, Ghosh, S. | | Deposit date: | 2019-12-24 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.713 Å) | | Cite: | Mycobacterial DNA polymerase I: activities and crystal structures of the POL domain as apoenzyme and in complex with a DNA primer-template and of the full-length FEN/EXO-POL enzyme.
Nucleic Acids Res., 48, 2020
|
|
6VDC
 
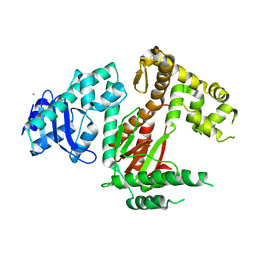 | | POL domain of Pol1 from M. smegmatis | | Descriptor: | DNA polymerase I, MANGANESE (II) ION | | Authors: | Shuman, S, Goldgur, Y, Ghosh, S. | | Deposit date: | 2019-12-24 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Mycobacterial DNA polymerase I: activities and crystal structures of the POL domain as apoenzyme and in complex with a DNA primer-template and of the full-length FEN/EXO-POL enzyme.
Nucleic Acids Res., 48, 2020
|
|
6VDD
 
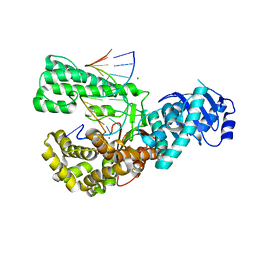 | | POL domain of Pol1 from M. smegmatis complex with DNA primer-template and dNTP | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, DNA (5'-D(*GP*CP*GP*AP*TP*CP*AP*CP*GP*TP*A*(DCT))-3'), DNA (5'-D(P*CP*GP*TP*AP*CP*GP*TP*GP*AP*TP*CP*GP*CP*A)-3'), ... | | Authors: | Shuman, S, Goldgur, Y, Ghosh, S. | | Deposit date: | 2019-12-24 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mycobacterial DNA polymerase I: activities and crystal structures of the POL domain as apoenzyme and in complex with a DNA primer-template and of the full-length FEN/EXO-POL enzyme.
Nucleic Acids Res., 48, 2020
|
|
4O8W
 
 | | Crystal Structure of the GerD spore germination protein | | Descriptor: | Spore germination protein | | Authors: | Li, Y, Jin, K, Ghosh, S, Devarakonda, P, Carlson, K, Davis, A, Stewart, K, Cammett, E, Rossi, P.P, Setlow, B, Lu, M, Setlow, P, Hao, B. | | Deposit date: | 2013-12-30 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.293 Å) | | Cite: | Structural and Functional Analysis of the GerD Spore Germination Protein of Bacillus Species.
J.Mol.Biol., 426, 2014
|
|
6EWN
 
 | | HspA from Thermosynechococcus vulcanus in the presence of 2M urea with initial stages of denaturation | | Descriptor: | HspA, UREA | | Authors: | Adir, N, Ghosh, S, Salama, F, Dines, M. | | Deposit date: | 2017-11-06 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Biophysical and structural characterization of the small heat shock protein HspA from Thermosynechococcus vulcanus in 2 M urea.
Biochim Biophys Acta Proteins Proteom, 1867, 2019
|
|
3TTJ
 
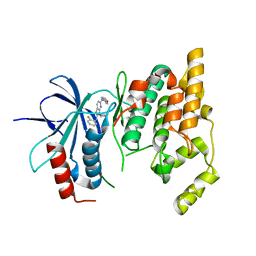 | | Crystal Structure of JNK3 complexed with CC-359, a JNK inhibitor for the prevention of ischemia-reperfusion injury | | Descriptor: | 9-cyclopentyl-N~8~-(2-fluorophenyl)-N~2~-(4-methoxyphenyl)-9H-purine-2,8-diamine, Mitogen-activated protein kinase 10 | | Authors: | Plantevin-Krenitsky, V, Delgado, M, Nadolny, L, Sahasrabudhe, K, Ayala, S, Clareen, S, Hilgraf, R, Albers, R, Kois, A, Hughes, K, Wright, J, Nowakowski, J, Sudbeck, E, Ghosh, S, Bahmanyar, S, Chamberlain, P, Muir, J, Cathers, B.E, Giegel, D, Xu, L, Celeridad, M, Moghaddam, M, Khatsenko, O, Omholt, P, Katz, J, Pai, S, Fan, R, Tang, Y, Shirley, M.A, Benish, B, Blease, K, Raymon, H, Bhagwat, S, Bennett, B, Satoh, Y. | | Deposit date: | 2011-09-14 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Aminopurine based JNK inhibitors for the prevention of ischemia reperfusion injury.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
1FN0
 
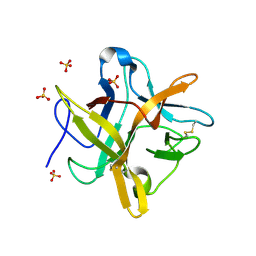 | | STRUCTURE OF A MUTANT WINGED BEAN CHYMOTRYPSIN INHIBITOR PROTEIN, N14D. | | Descriptor: | CHYMOTRYPSIN INHIBITOR 3, SULFATE ION | | Authors: | Dattagupta, J.K, Chakrabarti, C, Ravichandran, S, Dasgupta, J, Ghosh, S. | | Deposit date: | 2000-08-19 | | Release date: | 2001-02-19 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The role of Asn14 in the stability and conformation of the reactive-site loop of winged bean chymotrypsin inhibitor: crystal structures of two point mutants Asn14-->Lys and Asn14-->Asp.
PROTEIN ENG., 14, 2001
|
|
1FMZ
 
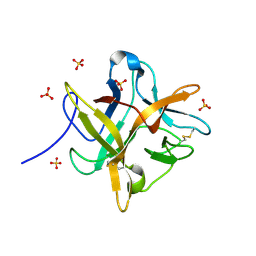 | | CRYSTAL STRUCTURE OF A MUTANT WINGED BEAN CHYMOTRYPSIN INHIBITOR PROTEIN, N14K. | | Descriptor: | CHYMOTRYPSIN INHIBITOR 3, SULFATE ION | | Authors: | Dattagupta, J.K, Chakrabarti, C, Ravichandran, S, Dasgupta, J, Ghosh, S. | | Deposit date: | 2000-08-19 | | Release date: | 2001-02-19 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The role of Asn14 in the stability and conformation of the reactive-site loop of winged bean chymotrypsin inhibitor: crystal structures of two point mutants Asn14-->Lys and Asn14-->Asp.
PROTEIN ENG., 14, 2001
|
|
1EYL
 
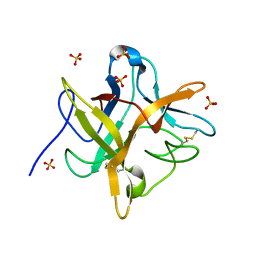 | | STRUCTURE OF A RECOMBINANT WINGED BEAN CHYMOTRYPSIN INHIBITOR | | Descriptor: | CHYMOTRYPSIN INHIBITOR, SULFATE ION | | Authors: | Dattagupta, J.K, Chakrabarti, C, Ravichandran, S, Ghosh, S. | | Deposit date: | 2000-05-07 | | Release date: | 2000-05-24 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The role of Asn14 in the stability and conformation of the reactive-site loop of winged bean chymotrypsin inhibitor: crystal structures of two point mutants Asn14-->Lys and Asn14-->Asp.
Protein Eng., 14, 2001
|
|
3Q0G
 
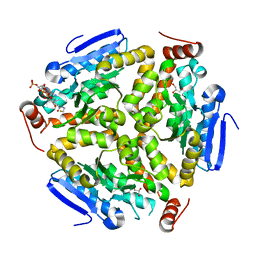 | | Crystal Structure of the Mycobacterium tuberculosis Crotonase Bound to a Reaction Intermediate Derived from Crotonyl CoA | | Descriptor: | Butyryl Coenzyme A, COENZYME A, GLYCEROL, ... | | Authors: | Bruning, J.B, Delgado, E, Ghosh, S, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-12-15 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal Structure of the Prokaryotic Crotonase
To be Published
|
|
3Q0J
 
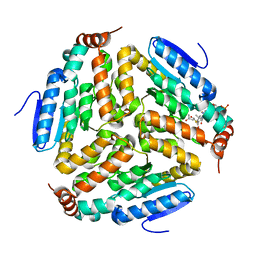 | |
3PZK
 
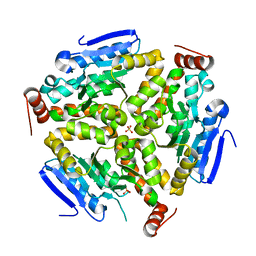 | |
3QRF
 
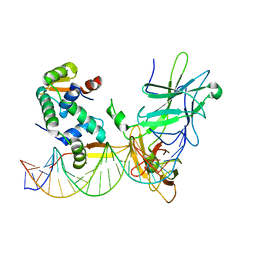 | | Structure of a domain-swapped FOXP3 dimer | | Descriptor: | Forkhead box protein P3, MAGNESIUM ION, Nuclear factor of activated T-cells, ... | | Authors: | Bandukwala, H.S, Wu, Y, Feurer, M, Chen, Y, Barbosa, B, Ghosh, S, Stroud, J.C, Benoist, C, Mathis, D, Rao, A, Chen, L. | | Deposit date: | 2011-02-17 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a Domain-Swapped FOXP3 Dimer on DNA and Its Function in Regulatory T Cells.
Immunity, 34, 2011
|
|
7VNL
 
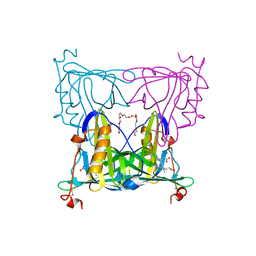 | | Sandercyanin mutant-F55A-Biliverdin complex | | Descriptor: | BILIVERDINE IX ALPHA, Sandercyanin Fluorescent Protein, TETRAETHYLENE GLYCOL | | Authors: | Yadav, K, Ghosh, S, Subramanian, R. | | Deposit date: | 2021-10-11 | | Release date: | 2022-05-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Phenylalanine stacking enhances the red fluorescence of biliverdin IX alpha on UV excitation in sandercyanin fluorescent protein.
Febs Lett., 596, 2022
|
|
7VNS
 
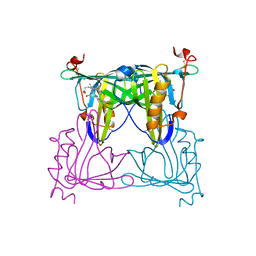 | | Sandercyanin mutant E79A-Biliverdin complex | | Descriptor: | BILIVERDINE IX ALPHA, Sandercyanin Fluorescent Protein | | Authors: | Yadav, K, Ghosh, S, Subramanian, R. | | Deposit date: | 2021-10-12 | | Release date: | 2022-05-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Phenylalanine stacking enhances the red fluorescence of biliverdin IX alpha on UV excitation in sandercyanin fluorescent protein.
Febs Lett., 596, 2022
|
|
3UE8
 
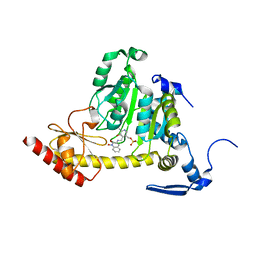 | | Kynurenine Aminotransferase II Inhibitors | | Descriptor: | (5-hydroxy-4-{[(1-hydroxy-2-oxo-1,2-dihydroquinolin-3-yl)amino]methyl}-6-methylpyridin-3-yl)methyl dihydrogen phosphate, CHLORIDE ION, Kynurenine/alpha-aminoadipate aminotransferase, ... | | Authors: | Dounay, A.B, Anderson, M, Bechle, B.M, Campbell, B.M, Claffey, M.M, Evdokimov, A, Edelweiss, E, Fonseca, K.R, Gan, X, Ghosh, S, Hayward, M.M, Horner, W, Kim, J.Y, McAllister, L.A, Pandit, J, Paradis, V, Parikh, V.D, Reese, M.R, Rong, S.B, Salafia, M.A, Schuyten, K, Strick, C.A, Tuttle, J.B, Valentine, J, Wang, H, Zawadzke, L.E, Verhoest, P.R. | | Deposit date: | 2011-10-28 | | Release date: | 2012-02-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Discovery of Brain-Penetrant, Irreversible Kynurenine Aminotransferase II Inhibitors for Schizophrenia.
ACS Med Chem Lett, 3, 2012
|
|
4M9F
 
 | | Dengue virus NS2B-NS3 protease A125C variant at pH 8.5 | | Descriptor: | NS2B-NS3 protease | | Authors: | Yildiz, M, Ghosh, S, Bell, J.A, Sherman, W, Hardy, J.A. | | Deposit date: | 2013-08-14 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Allosteric Inhibition of the NS2B-NS3 Protease from Dengue Virus.
Acs Chem.Biol., 8, 2013
|
|
2M6O
 
 | | The actinobacterial transcription factor RbpA binds to the principal sigma subunit of RNA polymerase | | Descriptor: | Uncharacterized protein | | Authors: | Liu, B, Tabib-Salazar, A, Doughty, P, Lewis, R, Ghosh, S, Parsy, M, Simpson, P, Matthews, S, Paget, M. | | Deposit date: | 2013-04-06 | | Release date: | 2013-05-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The actinobacterial transcription factor RbpA binds to the principal sigma subunit of RNA polymerase.
Nucleic Acids Res., 41, 2013
|
|
1Q54
 
 | | STRUCTURE AND MECHANISM OF ACTION OF ISOPENTENYLPYROPHOSPHATE-DIMETHYLALLYLPYROPHOSPHATE ISOMERASE: COMPLEX WITH THE BROMOHYDRINE OF IPP | | Descriptor: | 4-BROMO-3-HYDROXY-3-METHYL BUTYL DIPHOSPHATE, ISOPENTENYL DIPHOSPHATE DELTA-ISOMERASE, MAGNESIUM ION, ... | | Authors: | Wouters, J, Oudjama, Y, Ghosh, S, Stalon, V, Droogmans, L. | | Deposit date: | 2003-08-06 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure and mechanism of action of isopentenylpyrophosphate-dimethylallylpyrophosphate isomerase.
J.Am.Chem.Soc., 125, 2003
|
|
