4XPZ
 
 | |
4XQ0
 
 | |
7Z9L
 
 | | Phen-DC3 intercalation causes hybrid-to-antiparallel transformation of human telomeric DNA G-quadruplex | | Descriptor: | DNA (5'-D(*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*G)-3'), N2,N9-bis(1-methylquinolin-3-yl)-1,10-phenanthroline-2,9-dicarboxamide | | Authors: | Ghosh, A, Trajkovski, M, Teulade-Fichou, M.P, Gabelica, V, Plavec, J. | | Deposit date: | 2022-03-21 | | Release date: | 2022-08-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Phen-DC 3 Induces Refolding of Human Telomeric DNA into a Chair-Type Antiparallel G-Quadruplex through Ligand Intercalation.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
6E20
 
 | | Crystal structure of the Dario rerio galectin-1-L2 | | Descriptor: | Galectin, MAGNESIUM ION, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Ghosh, A, Bianchet, M.A. | | Deposit date: | 2018-07-10 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the zebrafish galectin-1-L2 and model of its interaction with the infectious hematopoietic necrosis virus (IHNV) envelope glycoprotein.
Glycobiology, 29, 2019
|
|
4RP4
 
 | |
4RP3
 
 | | Crystal Structure of the L27 Domain of Discs Large 1 (target ID NYSGRC-010766) from Drosophila melanogaster bound to a potassium ion (space group P212121) | | Descriptor: | CHLORIDE ION, Disks large 1 tumor suppressor protein, FORMIC ACID, ... | | Authors: | Ghosh, A, Ramagopal, U, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-10-29 | | Release date: | 2014-11-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structures of the L27 Domain of Disc Large Homologue 1 Protein Illustrate a Self-Assembly Module.
Biochemistry, 57, 2018
|
|
4RQB
 
 | |
4RQA
 
 | |
4RP5
 
 | |
2ALV
 
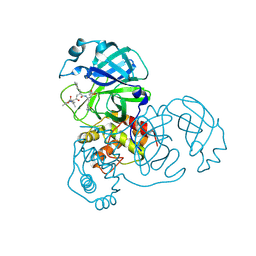 | | X-ray structural analysis of SARS coronavirus 3CL proteinase in complex with designed anti-viral inhibitors | | Descriptor: | N-((3S,6R)-6-((S,E)-4-ETHOXYCARBONYL-1-((S)-2-OXOPYRROLIDIN-3-YL)BUT-3-EN-2-YLCARBAMOYL)-2,9-DIMETHYL-4-OXODEC-8-EN-3-YL)-5-METHYLISOXAZOLE-3-CARBOXAMIDE, Replicase polyprotein 1ab | | Authors: | Ghosh, A.K, Xi, K, Ratia, K, Santarsiero, B.D, Fu, W, Harcourt, B.H, Rota, P.A, Baker, S.C, Johnson, M.E, Mesecar, A.D. | | Deposit date: | 2005-08-08 | | Release date: | 2006-08-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design and synthesis of peptidomimetic severe acute respiratory syndrome chymotrypsin-like protease inhibitors.
J.Med.Chem., 48, 2005
|
|
2D4H
 
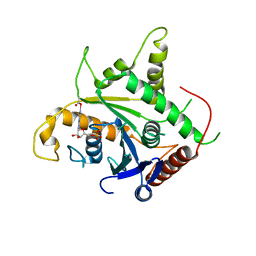 | | Crystal-structure of the N-terminal large GTPase Domain of human Guanylate Binding protein 1 (hGBP1) in complex with GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Interferon-induced guanylate-binding protein 1 | | Authors: | Ghosh, A, Praefcke, G.J.K, Renault, L, Wittinghofer, A, Herrmann, C. | | Deposit date: | 2005-10-19 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | How guanylate-binding proteins achieve assembly-stimulated processive cleavage of GTP to GMP.
Nature, 440, 2006
|
|
2B8W
 
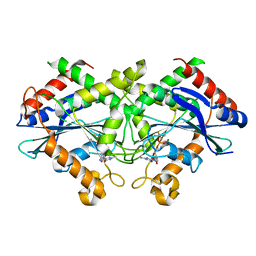 | | Crystal-structure of the N-terminal Large GTPase Domain of human Guanylate Binding protein 1 (hGBP1) in complex with GMP/AlF4 | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Interferon-induced guanylate-binding protein 1, MAGNESIUM ION, ... | | Authors: | Ghosh, A, Praefcke, G.J.K, Renault, L, Wittinghofer, A, Herrmann, C. | | Deposit date: | 2005-10-10 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | How guanylate-binding proteins achieve assembly-stimulated processive cleavage of GTP to GMP.
Nature, 440, 2006
|
|
2B92
 
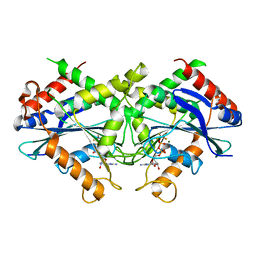 | | Crystal-structure of the N-terminal Large GTPase Domain of human Guanylate Binding protein 1 (hGBP1) in complex with GDP/AlF3 | | Descriptor: | ALUMINUM FLUORIDE, GUANOSINE-5'-DIPHOSPHATE, Interferon-induced guanylate-binding protein 1, ... | | Authors: | Ghosh, A, Praefcke, G.J.K, Renault, L, Wittinghofer, A, Herrmann, C. | | Deposit date: | 2005-10-10 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | How guanylate-binding proteins achieve assembly-stimulated processive cleavage of GTP to GMP.
Nature, 440, 2006
|
|
2BC9
 
 | | Crystal-structure of the N-terminal large GTPase Domain of human Guanylate Binding protein 1 (hGBP1) in complex with non-hydrolysable GTP analogue GppNHp | | Descriptor: | Interferon-induced guanylate-binding protein 1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Ghosh, A, Praefcke, G.J.K, Renault, L, Wittinghofer, A, Herrmann, C. | | Deposit date: | 2005-10-19 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | How guanylate-binding proteins achieve assembly-stimulated processive cleavage of GTP to GMP.
Nature, 440, 2006
|
|
2JR6
 
 | | Solution structure of UPF0434 protein NMA0874. Northeast Structural Genomics Target MR32 | | Descriptor: | UPF0434 protein NMA0874 | | Authors: | Ghosh, A, Singarapu, K.K, Wu, Y, Liu, G, Sukumaran, D, Chen, C.X, Nwosu, C, Owens, L, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-20 | | Release date: | 2007-07-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of UPF0434 protein NMA0874.
To be Published
|
|
1TPZ
 
 | | Crystal Structure of IIGP1: a paradigm for interferon inducible p47 resistance GTPases | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Ghosh, A, Uthaiah, R, Howard, J, Herrmann, C, Wolf, E. | | Deposit date: | 2004-06-16 | | Release date: | 2004-09-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of IIGP1; A Paradigm for Interferon-Inducible p47 Resistance GTPases
Mol.Cell, 15, 2004
|
|
1TQ4
 
 | | Crystal Structure of IIGP1: a paradigm for interferon inducible p47 resistance GTPases | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, interferon-inducible GTPase | | Authors: | Ghosh, A, Uthaiah, R, Howard, J, Herrmann, C, Wolf, E. | | Deposit date: | 2004-06-16 | | Release date: | 2004-09-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of IIGP1; A Paradigm for Interferon-Inducible p47 Resistance GTPases
Mol.Cell, 15, 2004
|
|
8UFN
 
 | |
8UFO
 
 | |
1TQ2
 
 | | Crystal Structure of IIGP1: a paradigm for interferon inducible p47 resistance GTPases | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Ghosh, A, Uthaiah, R, Howard, J, Herrmann, C, Wolf, E. | | Deposit date: | 2004-06-16 | | Release date: | 2004-09-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of IIGP1; A Paradigm for Interferon-Inducible p47 Resistance GTPases
Mol.Cell, 15, 2004
|
|
1TQ6
 
 | | Crystal Structure of IIGP1: a paradigm for interferon inducible p47 resistance GTPases | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, interferon-inducible GTPase | | Authors: | Ghosh, A, Uthaiah, R, Howard, J, Herrmann, C, Wolf, E. | | Deposit date: | 2004-06-16 | | Release date: | 2004-09-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of IIGP1; A Paradigm for Interferon-Inducible p47 Resistance GTPases
Mol.Cell, 15, 2004
|
|
1TQD
 
 | | Crystal structure of IIGP1: a paradigm for interferon inducible p47 resistance GTPases | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SULFATE ION, interferon-inducible GTPase | | Authors: | Ghosh, A, Uthaiah, R, Howard, J, Herrmann, C, Wolf, E. | | Deposit date: | 2004-06-17 | | Release date: | 2004-09-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of IIGP1; A Paradigm for Interferon-Inducible p47 Resistance GTPases
Mol.Cell, 15, 2004
|
|
4GID
 
 | | Structure of beta-secretase complexed with inhibitor | | Descriptor: | Beta-secretase 1, L-PROLINAMIDE, N-[(2S)-1-({(2S,3R)-3-hydroxy-1-[(2-methylpropyl)amino]-1-oxobutan-2-yl}amino)-3-phenylpropan-2-yl]-5-[methyl(methylsulfonyl)amino]-N'-[(1R)-1-phenylethyl]benzene-1,3-dicarboxamide | | Authors: | Ghosh, A, Tang, J, Venkateswara, R.K, Yadav, N, Anderson, D, Gavande, N, Huang, X, Terzyan, S. | | Deposit date: | 2012-08-08 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design of highly selective beta-secretase inhibitors: synthesis, biological evaluation, and protein-ligand X-ray crystal structure.
J.Med.Chem., 55, 2012
|
|
3EF0
 
 | |
3VC7
 
 | | Crystal Structure of a putative oxidoreductase from Sinorhizobium meliloti 1021 | | Descriptor: | GLYCEROL, Putative oxidoreductase | | Authors: | Ghosh, A, Bhoshle, R, Toro, R, Gizzi, A, Hillerich, B, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-01-03 | | Release date: | 2012-01-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.231 Å) | | Cite: | Crystal Structure of a putative oxidoreductase protein from Sinorhizobium meliloti 1021
To be Published
|
|
