8SWR
 
 | | Structure of K. lactis PNP S42E variant bound to transition state analog DADMe-IMMUCILLIN G and sulfate | | 分子名称: | 2-amino-7-{[(3R,4R)-3-hydroxy-4-(hydroxymethyl)pyrrolidin-1-yl]methyl}-3,5-dihydro-4H-pyrrolo[3,2-d]pyrimidin-4-one, GUANINE, Purine nucleoside phosphorylase, ... | | 著者 | Fedorov, E, Ghosh, A. | | 登録日 | 2023-05-19 | | 公開日 | 2023-10-18 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Phosphate Binding in PNP Alters Transition-State Analogue Affinity and Subunit Cooperativity.
Biochemistry, 62, 2023
|
|
8SWQ
 
 | | Structure of K. lactis PNP bound to transition state analog DADMe-IMMUCILLIN H and sulfate | | 分子名称: | 7-[[(3R,4R)-3-(hydroxymethyl)-4-oxidanyl-pyrrolidin-1-ium-1-yl]methyl]-3,5-dihydropyrrolo[3,2-d]pyrimidin-4-one, GLYCEROL, Purine nucleoside phosphorylase, ... | | 著者 | Fedorov, E, Ghosh, A. | | 登録日 | 2023-05-19 | | 公開日 | 2023-10-18 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.979 Å) | | 主引用文献 | Phosphate Binding in PNP Alters Transition-State Analogue Affinity and Subunit Cooperativity.
Biochemistry, 62, 2023
|
|
8SWT
 
 | |
8FUI
 
 | | HIV-1 wild type protease with GRL-02519A, with N-(2,5-dimethylphenyl)-4-(pyridin-3-yl)pyrimidin-2-amine as P2-P3 group | | 分子名称: | ACETATE ION, CHLORIDE ION, FORMIC ACID, ... | | 著者 | Wang, Y.-F, Wong-Sam, A.E, Ghosh, A.K, Weber, I.T. | | 登録日 | 2023-01-17 | | 公開日 | 2023-05-24 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Exploration of imatinib and nilotinib-derived templates as the P2-Ligand for HIV-1 protease inhibitors: Design, synthesis, protein X-ray structural studies, and biological evaluation.
Eur.J.Med.Chem., 255, 2023
|
|
8FUJ
 
 | | HIV-1 wild type protease with GRL-03419A, with N-(2,5-dimethylphenyl)-4-(pyridin-3-yl)pyrimidin-2-amine as P2-P3 group and 3,5-difluorophenylmethyl as the P1 group | | 分子名称: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | 著者 | Wang, Y.-F, Agniswamy, J, Ghosh, A.K, Weber, I.T. | | 登録日 | 2023-01-17 | | 公開日 | 2023-05-24 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.12 Å) | | 主引用文献 | Exploration of imatinib and nilotinib-derived templates as the P2-Ligand for HIV-1 protease inhibitors: Design, synthesis, protein X-ray structural studies, and biological evaluation.
Eur.J.Med.Chem., 255, 2023
|
|
8SWS
 
 | | Structure of K. lactis PNP S42E-H98R variant bound to transition state analog DADMe-IMMUCILLIN G and sulfate | | 分子名称: | 2-amino-7-{[(3R,4R)-3-hydroxy-4-(hydroxymethyl)pyrrolidin-1-yl]methyl}-3,5-dihydro-4H-pyrrolo[3,2-d]pyrimidin-4-one, Purine nucleoside phosphorylase, SULFATE ION | | 著者 | Fedorov, E, Ghosh, A. | | 登録日 | 2023-05-19 | | 公開日 | 2023-10-18 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Phosphate Binding in PNP Alters Transition-State Analogue Affinity and Subunit Cooperativity.
Biochemistry, 62, 2023
|
|
8SWU
 
 | |
6OGR
 
 | | X-ray crystal structure of darunavir-resistant HIV-1 protease (P30) in complex with GRL-142 | | 分子名称: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3,5-difluorophenyl)-3-hydroxybutan-2-yl]carbamate, 1,2-ETHANEDIOL, Protease | | 著者 | Bulut, H, Hattori, S.I, Aoki-Ogata, H, Hayashi, H, Aoki, M, Ghosh, A.K, Mitsuya, H. | | 登録日 | 2019-04-03 | | 公開日 | 2020-04-08 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.28 Å) | | 主引用文献 | Novel HIV-1 protease inhibitors, GRL-142 and its analogs, markedly adapt to the structural plasticity of HIV-1 protease and exerts extreme potency with high genetic barrier
To Be Published
|
|
6OTN
 
 | | Crystal Structure of an N-terminal Fragment of Cancer Associated Tropomyosin 3.1 (Tpm3.1) | | 分子名称: | SULFATE ION, Tropomyosin alpha-3 chain | | 著者 | Rynkiewicz, M.J, Ghosh, A, Lehman, W.J, Janco, M, Gunning, P.W. | | 登録日 | 2019-05-03 | | 公開日 | 2019-08-14 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Molecular integration of the anti-tropomyosin compound ATM-3507 into the coiled coil overlap region of the cancer-associated Tpm3.1.
Sci Rep, 9, 2019
|
|
6XR3
 
 | |
8GAC
 
 | | Crystal structure of a high affinity CTLA-4 binder | | 分子名称: | 1,2-ETHANEDIOL, CTLA-4 binder | | 著者 | Yang, W, Almo, S.C, Baker, D, Ghosh, A. | | 登録日 | 2023-02-22 | | 公開日 | 2024-08-21 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Design of High Affinity Binders to Convex Protein Target Sites.
Biorxiv, 2024
|
|
7L1A
 
 | | Human Methionine Adenosyltransferase 2A bound to Methylthioadenosine and inhibitor, di-imido triphosphate (PNPNP) | | 分子名称: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, ALANINE, MAGNESIUM ION, ... | | 著者 | Niland, C.N, Fedorov, E, Schramm, V.L, Ghosh, A. | | 登録日 | 2020-12-14 | | 公開日 | 2021-03-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Mechanism and Inhibition of Human Methionine Adenosyltransferase 2A.
Biochemistry, 60, 2021
|
|
8GAD
 
 | |
8GAB
 
 | | Crystal structure of CTLA-4 in complex with a high affinity CTLA-4 binder | | 分子名称: | CTLA-4 binder, Cytotoxic T-lymphocyte protein 4, POTASSIUM ION | | 著者 | Yang, W, Almo, S.C, Baker, D, Ghosh, A. | | 登録日 | 2023-02-22 | | 公開日 | 2024-08-21 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.72 Å) | | 主引用文献 | Design of High Affinity Binders to Convex Protein Target Sites.
Biorxiv, 2024
|
|
9C66
 
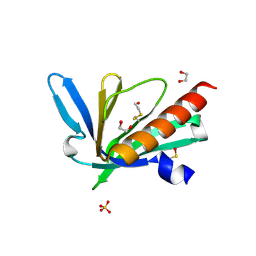 | | Structure of the Mena EVH1 domain bound to the polyproline segment of PTP1B | | 分子名称: | 1,2-ETHANEDIOL, Protein enabled homolog, SULFATE ION, ... | | 著者 | LaComb, L, Fedorov, E, Bonanno, J.B, Almo, S.C, Ghosh, A. | | 登録日 | 2024-06-07 | | 公開日 | 2024-08-28 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Insights into the Interaction Landscape of the EVH1 Domain of Mena.
Biochemistry, 63, 2024
|
|
9C6V
 
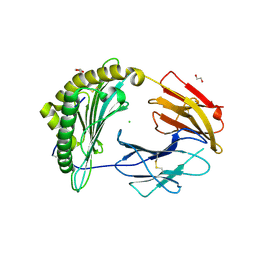 | | Crystal Structure of a single chain trimer composed of HLA-B*39:06 Y84C variant, beta-2microglobulin, and NRVMLPKAA peptide from NLRP2 (1 molecule/asymmetric unit) | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | 著者 | Sharma, R, Amdare, N.P, Celikgil, A, Garforth, S.J, DiLorenzo, T.P, Almo, S.C, Ghosh, A. | | 登録日 | 2024-06-09 | | 公開日 | 2024-09-04 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural and biochemical analysis of highly similar HLA-B allotypes differentially associated with type 1 diabetes.
J.Biol.Chem., 300, 2024
|
|
9C6X
 
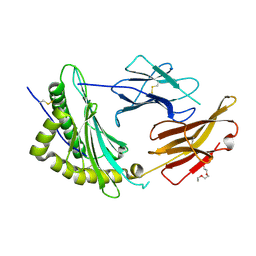 | | Crystal Structure of a single chain trimer composed of HLA-B*39:01 Y84C variant, beta-2microglobulin, and NRVMLPKAA peptide from NLRP2 | | 分子名称: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, NACHT, ... | | 著者 | Sharma, R, Amdare, N.P, Celikgil, A, Garforth, S.J, DiLorenzo, T.P, Almo, S.C, Ghosh, A. | | 登録日 | 2024-06-09 | | 公開日 | 2024-09-04 | | 最終更新日 | 2024-10-02 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural and biochemical analysis of highly similar HLA-B allotypes differentially associated with type 1 diabetes.
J.Biol.Chem., 300, 2024
|
|
9C6W
 
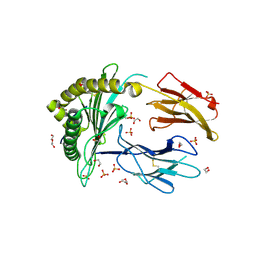 | | Crystal Structure of a single chain trimer composed of HLA-B*39:06 Y84C variant, beta-2microglobulin, and NRVMLPKAA peptide from NLRP2 (2 molecules/asymmetric unit) | | 分子名称: | 1,2-ETHANEDIOL, 3,6,9,12,15-PENTAOXAHEPTADECANE, CHLORIDE ION, ... | | 著者 | Sharma, R, Amdare, N.P, Celikgil, A, Garforth, S.J, DiLorenzo, T.P, Almo, S.C, Ghosh, A. | | 登録日 | 2024-06-09 | | 公開日 | 2024-09-04 | | 最終更新日 | 2024-10-02 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural and biochemical analysis of highly similar HLA-B allotypes differentially associated with type 1 diabetes.
J.Biol.Chem., 300, 2024
|
|
7JPJ
 
 | | Crystal Structure of the essential dimeric LYSA from Phaeodactylum tricornutum | | 分子名称: | D-LYSINE, Diaminopimelate decarboxylase, SULFATE ION | | 著者 | Fedorov, E, Belinski, V.A, Brunson, J.K, Almo, S.C, Dupont, C.L, Ghosh, A. | | 登録日 | 2020-08-08 | | 公開日 | 2021-06-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.78 Å) | | 主引用文献 | The Phaeodactylum tricornutum diaminopimelate decarboxylase was acquired via horizontal gene transfer from bacteria and displays substrate promiscuity
Biorxiv, 2020
|
|
6UWC
 
 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-08613 | | 分子名称: | (3aS,4S,7aR)-hexahydro-4H-furo[2,3-b]pyran-4-yl {(2S,3R)-1-(4-fluorophenyl)-3-hydroxy-4-[(2-methylpropyl)({2-[(propan-2-yl)amino]-1,3-benzothiazol-6-yl}sulfonyl)amino]butan-2-yl}carbamate, Protease | | 著者 | Yedidi, R.S, Hayashi, H, Ghosh, A.K, Mitsuya, H. | | 登録日 | 2019-11-05 | | 公開日 | 2020-11-18 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Fluorine Modifications Contribute to Potent Antiviral Activity against Highly Drug-Resistant HIV-1 and Favorable Blood-Brain Barrier Penetration Property of Novel Central Nervous System-Targeting HIV-1 Protease Inhibitors In Vitro.
Antimicrob.Agents Chemother., 66, 2022
|
|
6UWB
 
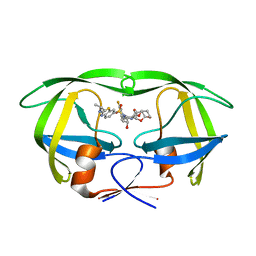 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-08513 | | 分子名称: | (3aS,4S,7aR)-hexahydro-4H-furo[2,3-b]pyran-4-yl {(2S,3R)-1-(3,5-difluorophenyl)-3-hydroxy-4-[(2-methylpropyl)({2-[(propan-2-yl)amino]-1,3-benzothiazol-6-yl}sulfonyl)amino]butan-2-yl}carbamate, Protease | | 著者 | Yedidi, R.S, Hayashi, H, Ghosh, A.K, Mitsuya, H. | | 登録日 | 2019-11-05 | | 公開日 | 2020-11-18 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Fluorine Modifications Contribute to Potent Antiviral Activity against Highly Drug-Resistant HIV-1 and Favorable Blood-Brain Barrier Penetration Property of Novel Central Nervous System-Targeting HIV-1 Protease Inhibitors In Vitro.
Antimicrob.Agents Chemother., 66, 2022
|
|
5KAO
 
 | | Crystal structure of wild type HIV-1 protease in complex with GRL-10413 | | 分子名称: | [(3~{a}~{S},4~{R},6~{a}~{R})-2,3,3~{a},4,5,6~{a}-hexahydrofuro[2,3-b]furan-4-yl] ~{N}-[(2~{S},3~{R})-1-(3-chloranyl-4-methoxy-phenyl)-4-[(4-methoxyphenyl)sulfonyl-(2-methylpropyl)amino]-3-oxidanyl-butan-2-yl]carbamate, protease | | 著者 | Yedidi, R.S, Delino, N.S, Das, D, Kaufman, J.D, Wingfield, P.T, Ghosh, A.K, Mitsuya, H. | | 登録日 | 2016-06-01 | | 公開日 | 2016-08-31 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | A Modified P1 Moiety Enhances In Vitro Antiviral Activity against Various Multidrug-Resistant HIV-1 Variants and In Vitro Central Nervous System Penetration Properties of a Novel Nonpeptidic Protease Inhibitor, GRL-10413.
Antimicrob.Agents Chemother., 60, 2016
|
|
6D0D
 
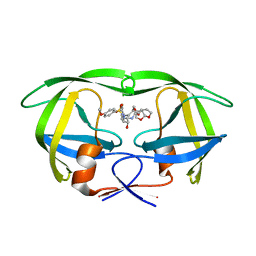 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-087-13 | | 分子名称: | (3aS,4S,7aR)-hexahydro-4H-furo[2,3-b]pyran-4-yl [(2S,3R)-1-(4-fluorophenyl)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}butan-2-yl]carbamate, Protease | | 著者 | Yedidi, R.S, Hayashi, H, Ghosh, A.K, Mitsuya, H. | | 登録日 | 2018-04-10 | | 公開日 | 2019-05-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Novel Central Nervous System (CNS)-Targeting Protease Inhibitors for Drug-Resistant HIV Infection and HIV-Associated CNS Complications.
Antimicrob.Agents Chemother., 63, 2019
|
|
6D0E
 
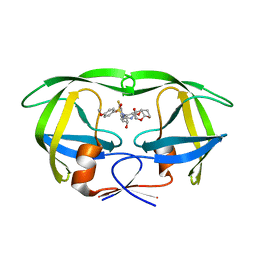 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-084-13 | | 分子名称: | (3aS,4S,7aR)-hexahydro-4H-furo[2,3-b]pyran-4-yl [(2S,3R)-1-(3,5-difluorophenyl)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}butan-2-yl]carbamate, Protease | | 著者 | Yedidi, R.S, Hayashi, H, Ghosh, A.K, Mitsuya, H. | | 登録日 | 2018-04-10 | | 公開日 | 2019-05-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Novel Central Nervous System (CNS)-Targeting Protease Inhibitors for Drug-Resistant HIV Infection and HIV-Associated CNS Complications.
Antimicrob.Agents Chemother., 63, 2019
|
|
2Z4O
 
 | | Wild Type HIV-1 Protease with potent Antiviral inhibitor GRL-98065 | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(2S,3R)-3-HYDROXY-4-(N-ISOBUTYLBENZO[D][1,3]DIOXOLE-5-SULFONAMIDO)-1-PHENYLBUTAN-2-YLCARBAMATE, ACETATE ION, CHLORIDE ION, ... | | 著者 | Wang, Y.F, Tie, Y, Boross, P.I, Tozser, J, Ghosh, A.K, Harrison, R.W, Weber, I.T. | | 登録日 | 2007-06-20 | | 公開日 | 2008-04-22 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Potent new antiviral compound shows similar inhibition and structural interactions with drug resistant mutants and wild type HIV-1 protease.
J.Med.Chem., 50, 2007
|
|
