3VCC
 
 | | CRYSTAL STRUCTURE OF D-Galacturonate Dehydratase from GEOBACILLUS SP. complexed with Mg | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, Mandelate racemase/muconate lactonizing protein | | Authors: | Fedorov, A.A, Fedorov, E.V, Groninger-Poe, F, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2012-01-03 | | Release date: | 2013-01-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | CRYSTAL STRUCTURE OF D-Galacturonate Dehydratase from GEOBACILLUS SP. complexed with Mg
To be Published
|
|
2QJM
 
 | | Crystal structure of the K271E mutant of Mannonate dehydratase from Novosphingobium aromaticivorans complexed with Mg and D-mannonate | | Descriptor: | D-MANNONIC ACID, MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme | | Authors: | Fedorov, A.A, Fedorov, E.V, Rakus, J.F, Vick, J.E, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2007-07-08 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: D-Mannonate dehydratase from Novosphingobium aromaticivorans.
Biochemistry, 46, 2007
|
|
4LUI
 
 | | Crystal structure of Orotidine 5'-monophosphate decarboxylase from methanocaldococcus jannaschii | | Descriptor: | CHLORIDE ION, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2013-07-25 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Orotidine 5'-monophosphate decarboxylase from methanocaldococcus jannaschii
To be Published
|
|
4MCO
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Rhodoferax ferrireducens (Rfer_1840), target EFI-510211, with bound malonate | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MALONATE ION, TRAP dicarboxylate transporter-DctP subunit | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Zhao, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-08-21 | | Release date: | 2013-09-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4MEV
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Rhodoferax ferrireducens (Rfer_1840), Target EFI-510211, with bound malonate, space group I422 | | Descriptor: | CITRIC ACID, MALONATE ION, TRAP dicarboxylate transporter-DctP subunit | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Zhao, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-08-27 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4LW7
 
 | | Crystal structure of the mutant H128S of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with the inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2013-07-26 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.423 Å) | | Cite: | Crystal structure of the mutant H128S of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with the inhibitor BMP
To be Published
|
|
4MF5
 
 | | Crystal structure of glutathione transferase BgramDRAFT_1843 from Burkholderia graminis, Target EFI-507289, with traces of one GSH bound | | Descriptor: | FORMIC ACID, GLUTATHIONE, Glutathione S-transferase domain | | Authors: | Patskovsky, Y, Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-08-27 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Crystal structure of glutathione transferase BgramDRAFT_1843 from Burkholderia graminis, Target EFI-507289, with traces of one GSH bound
To be Published
|
|
4MP4
 
 | | Crystal structure of a glutathione transferase family member from Acinetobacter baumannii, Target EFI-501785, apo structure | | Descriptor: | Glutathione S-transferase, SULFATE ION | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-09-12 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Crystal structure of a glutathione transferase family member from Acinetobacter baumannii, Target EFI-501785, apo structure
To be Published
|
|
4L8E
 
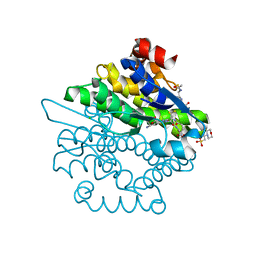 | | Crystal structure of a glutathione transferase family member from xenorhabdus nematophila, target efi-507418, with two gsh per subunit | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GLUTATHIONE, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-06-17 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a glutathione transferase family member from xenorhabdus nematophila, target efi-507418, with two gsh per subunit
To be published
|
|
4LB0
 
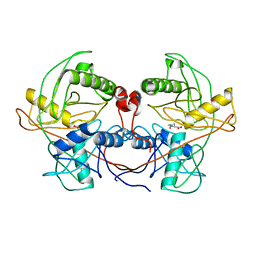 | | Crystal structure of a hydroxyproline epimerase from agrobacterium vitis, target efi-506420, with bound trans-4-oh-l-proline | | Descriptor: | 4-HYDROXYPROLINE, ACETATE ION, Uncharacterized protein | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Glenn, A.S, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-06-20 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a hydroxyproline epimerase from agrobacterium vitis, target efi-506420, with bound trans-4-oh-l-proline
To be Published
|
|
4LC6
 
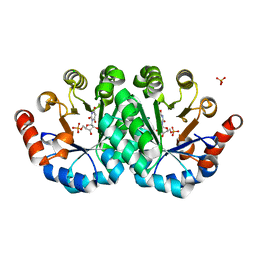 | | Crystal structure of the mutant H128Q of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with the inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, CHLORIDE ION, Orotidine 5'-phosphate decarboxylase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2013-06-21 | | Release date: | 2013-07-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Crystal structure of the mutant H128Q of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with the inhibitor BMP
To be Published
|
|
4LFE
 
 | | Crystal structure of geranylgeranyl diphosphate synthase sub1274 (target efi-509455) from streptococcus uberis 0140j with bound magnesium and isopentyl diphosphate, partially liganded complex; | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Geranylgeranyl diphosphate synthase, MAGNESIUM ION | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-06-26 | | Release date: | 2013-07-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Geranylgeranyl Diphosphate Synthase from Streptococcus Uberis 0140J
To be Published
|
|
4LLS
 
 | | Crystal structure of a farnesyl diphosphate synthase from Roseobacter denitrificans OCh 114, target EFI-509393, with IPP, GSPP, and calcium bound in active site | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, CALCIUM ION, GERANYL S-THIOLODIPHOSPHATE, ... | | Authors: | Kim, J, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Stead, M, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-07-09 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of a farnesyl diphosphate synthase from Roseobacter denitrificans OCh 114, target EFI-509393, with IPP, GSPP and calcium bound in active site
To be Published
|
|
4LFG
 
 | | Crystal structure of geranylgeranyl diphosphate synthase sub1274 (target efi-509455) from streptococcus uberis 0140j with bound magnesium and isopentyl diphosphate, fully liganded complex; | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Geranylgeranyl Diphosphate Synthase, MAGNESIUM ION | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-06-26 | | Release date: | 2013-07-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal Structure of Geranylgeranyl Diphosphate Synthase from Streptococcus Uberis 0140J
To be Published
|
|
4LLT
 
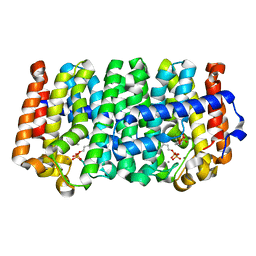 | | Crystal structure of a farnesyl diphosphate synthase from Roseobacter denitrificans OCh 114, target EFI-509393, with two IPP and calcium bound in active site | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, CALCIUM ION, Geranyltranstransferase | | Authors: | Kim, J, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Stead, M, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-07-09 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of a farnesyl diphosphate synthase from Roseobacter denitrificans OCh 114, target EFI-509393, with two IPP and calcium bound in active site
To be Published
|
|
4LN5
 
 | | Crystal structure of a trap periplasmic solute binding protein from burkholderia ambifaria (Bamb_6123), TARGET EFI-510059, with bound glycerol and chloride ion | | Descriptor: | CHLORIDE ION, GLYCEROL, TRAP dicarboxylate transporter, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-07-11 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4LOB
 
 | | Crystal structure of polyprenyl diphosphate synthase A1S_2732 (Target EFI-509223) from Acinetobacter baumannii | | Descriptor: | CHLORIDE ION, GLYCEROL, Polyprenyl synthetase | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-07-12 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of polyprenyl diphosphate synthase from Acinetobacter baumannii
To be Published
|
|
4NX5
 
 | | Crystal structure of orotidine 5'-monophosphate decarboxylase from methanobacterium thermoautotrophicum complexed with 6-azauridine 5'-monophosphate | | Descriptor: | 1,2-ETHANEDIOL, 6-AZA URIDINE 5'-MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Chan, K.K, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2013-12-08 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.591 Å) | | Cite: | Crystal structure of orotidine 5'-monophosphate decarboxylase from methanobacterium thermoautotrophicum complexed with 6-azauridine 5'-monophosphate
To be Published
|
|
4MHF
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Polaromonas sp. JS666 (Bpro_3107), target EFI-510173, with bound alpha/beta D-Glucuronate, space group P21 | | Descriptor: | TRAP dicarboxylate transporter, DctP subunit, alpha-D-glucopyranuronic acid, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Zhao, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-08-29 | | Release date: | 2013-09-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4NAP
 
 | | Crystal structure of a trap periplasmic solute binding protein from Desulfovibrio alaskensis G20 (DDE_0634), target EFI-510102, with bound d-tryptophan | | Descriptor: | D-TRYPTOPHAN, Extracellular solute-binding protein, family 7 | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-22 | | Release date: | 2013-11-13 | | Last modified: | 2015-02-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4N8G
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Chromohalobacter salexigens DSM 3043 (Csal_0660), Target EFI-501075, with bound D-alanine-D-alanine | | Descriptor: | CHLORIDE ION, D-ALANINE, SULFATE ION, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-17 | | Release date: | 2013-10-30 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4MZW
 
 | | CRYSTAL STRUCTURE OF NU-CLASS GLUTATHIONE TRANSFERASE YGHU FROM Streptococcus sanguinis SK36, COMPLEX WITH GLUTATHIONE DISULFIDE, TARGET EFI-507286 | | Descriptor: | ACETATE ION, Glutathione S-Transferase, OXIDIZED GLUTATHIONE DISULFIDE | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-09-30 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Glutathione S-Transferase Yghu (Target Efi-507286)
To be Published
|
|
4NGU
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Desulfovibrio alaskensis G20 (Dde_1548), Target EFI-510103, with bound D-Ala-D-Ala | | Descriptor: | CHLORIDE ION, D-ALANINE, SULFATE ION, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-11-02 | | Release date: | 2013-12-04 | | Last modified: | 2015-02-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4NT0
 
 | | Crystal structure of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with 3-deazauridine 5'-monophosphate | | Descriptor: | 4-hydroxy-1-(5-O-phosphono-beta-D-ribofuranosyl)pyridin-2(1H)-one, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Iiams, V, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2013-11-29 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.769 Å) | | Cite: | Crystal structure of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with 3-deazauridine 5'-monophosphate
To be Published
|
|
4LUJ
 
 | | Crystal structure of Orotidine 5'-monophosphate decarboxylase from methanocaldococcus jannaschii complexed with inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2013-07-25 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Orotidine 5'-monophosphate decarboxylase from methanocaldococcus jannaschii complexed with inhibitor BMP
To be Published
|
|
