3BEP
 
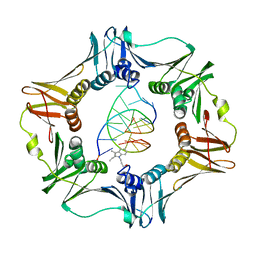 | | Structure of a sliding clamp on DNA | | 分子名称: | 1-(3-hydroxypropyl)-2-{(1E,3E,5E)-5-[1-(3-hydroxypropyl)-3,3-dimethyl-1,3-dihydro-2H-indol-2-ylidene]penta-1,3-dien-1-y l}-3,3-dimethyl-3H-indolium, DNA (5'-D(*DTP*DTP*DTP*DTP*DAP*DTP*DAP*DCP*DGP*DAP*DTP*DGP*DGP*DG)-3'), DNA (5'-D(P*DCP*DCP*DCP*DAP*DTP*DCP*DGP*DTP*DAP*DT)-3'), ... | | 著者 | Georgescu, R.E, Kim, S.S, Yurieva, O, Kuriyan, J, Kong, X.-P, O'Donnell, M. | | 登録日 | 2007-11-19 | | 公開日 | 2008-01-29 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Structure of a sliding clamp on DNA
Cell(Cambridge,Mass.), 132, 2008
|
|
3D1F
 
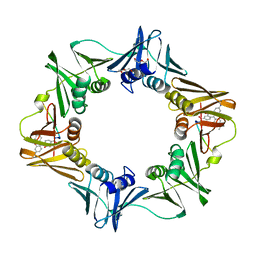 | | Crystal structure of E. coli sliding clamp (beta) bound to a polymerase III peptide | | 分子名称: | 2-[3,6-bis(dimethylamino)xanthen-9-yl]-5-methanoyl-benzoate, DI(HYDROXYETHYL)ETHER, DNA polymerase III subunit beta, ... | | 著者 | Georgescu, R.E, Yurieva, O, Seung-Sup, K, Kuriyan, J, Kong, X.-P, O'Donnell, M. | | 登録日 | 2008-05-05 | | 公開日 | 2008-07-29 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of a small-molecule inhibitor of a DNA polymerase sliding clamp.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3D1G
 
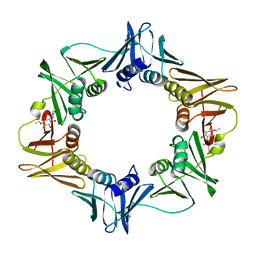 | | Structure of a small molecule inhibitor bound to a DNA sliding clamp | | 分子名称: | DNA polymerase III subunit beta, [(5R)-5-(2,3-dibromo-5-ethoxy-4-hydroxybenzyl)-4-oxo-2-thioxo-1,3-thiazolidin-3-yl]acetic acid | | 著者 | Georgescu, R.E, Yurieva, O, Seung-Sup, K, Kuriyan, J, Kong, X.-P, O'Donnell, M. | | 登録日 | 2008-05-05 | | 公開日 | 2008-07-29 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Structure of a small-molecule inhibitor of a DNA polymerase sliding clamp.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3D1E
 
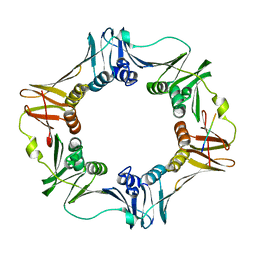 | | Crystal structure of E. coli sliding clamp (beta) bound to a polymerase II peptide | | 分子名称: | DNA polymerase III subunit beta, decamer from polymerase II C-terminal | | 著者 | Georgescu, R.E, Yurieva, O, Seung-Sup, K, Kuriyan, J, Kong, X.-P, O'Donnell, M. | | 登録日 | 2008-05-05 | | 公開日 | 2008-07-29 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure of a small-molecule inhibitor of a DNA polymerase sliding clamp.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
7SGZ
 
 | | Structure of the yeast Rad24-RFC loader bound to DNA and the closed 9-1-1 clamp | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, Crick strand, ... | | 著者 | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | 登録日 | 2021-10-07 | | 公開日 | 2022-03-23 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.17 Å) | | 主引用文献 | DNA is loaded through the 9-1-1 DNA checkpoint clamp in the opposite direction of the PCNA clamp.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7SH2
 
 | | Structure of the yeast Rad24-RFC loader bound to DNA and the open 9-1-1 clamp | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, Crick strand, ... | | 著者 | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | 登録日 | 2021-10-07 | | 公開日 | 2022-03-23 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.23 Å) | | 主引用文献 | DNA is loaded through the 9-1-1 DNA checkpoint clamp in the opposite direction of the PCNA clamp.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8THD
 
 | | Structure of the Saccharomyces cerevisiae clamp unloader Elg1-RFC bound to PCNA | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ELG1 isoform 1, MAGNESIUM ION, ... | | 著者 | Zheng, F, Yao, Y.N, Georgescu, R, O'Donnell, M.E, Li, H. | | 登録日 | 2023-07-14 | | 公開日 | 2024-05-22 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.25 Å) | | 主引用文献 | Structure of the PCNA unloader Elg1-RFC.
Sci Adv, 10, 2024
|
|
7KC0
 
 | | Structure of the Saccharomyces cerevisiae replicative polymerase delta in complex with a primer/template and the PCNA clamp | | 分子名称: | 2',3'-DIDEOXY-THYMIDINE-5'-TRIPHOSPHATE, DNA (25-MER), DNA (5'-D(P*AP*TP*GP*AP*CP*CP*AP*TP*GP*AP*TP*TP*AP*CP*GP*AP*AP*TP*TP*GP*C)-3'), ... | | 著者 | Zheng, F, Georgescu, R, Li, H, O'Donnell, M.E. | | 登録日 | 2020-10-04 | | 公開日 | 2020-12-02 | | 最終更新日 | 2020-12-16 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structure of eukaryotic DNA polymerase delta bound to the PCNA clamp while encircling DNA.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6WJV
 
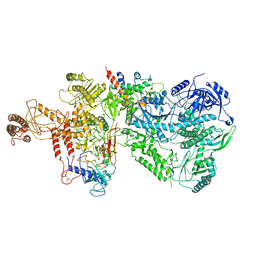 | | Structure of the Saccharomyces cerevisiae polymerase epsilon holoenzyme | | 分子名称: | DNA polymerase epsilon catalytic subunit A, DNA polymerase epsilon subunit B, DNA polymerase epsilon subunit C, ... | | 著者 | Yuan, Z, Georgescu, R, Schauer, G.D, O'Donnell, M, Li, H. | | 登録日 | 2020-04-14 | | 公開日 | 2020-07-08 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structure of the polymerase epsilon holoenzyme and atomic model of the leading strand replisome.
Nat Commun, 11, 2020
|
|
6U0M
 
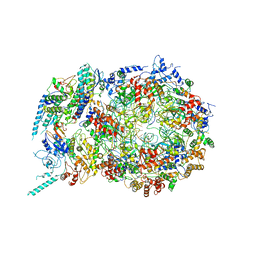 | | Structure of the S. cerevisiae replicative helicase CMG in complex with a forked DNA | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 45, DNA (15-MER), ... | | 著者 | Yuan, Z, Georgescu, R, Bai, L, Zhang, D, O'Donnell, M, Li, H. | | 登録日 | 2019-08-14 | | 公開日 | 2020-03-25 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | DNA unwinding mechanism of a eukaryotic replicative CMG helicase.
Nat Commun, 11, 2020
|
|
8THB
 
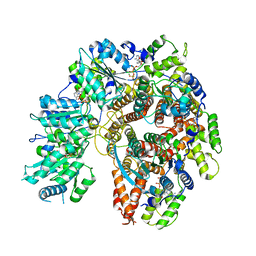 | | Structure of the Saccharomyces cerevisiae PCNA clamp unloader Elg1-RFC complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ELG1 isoform 1, MAGNESIUM ION, ... | | 著者 | Zheng, F, Yao, Y.N, Georgescu, R, O'Donnell, M.E, Li, H. | | 登録日 | 2023-07-14 | | 公開日 | 2024-05-22 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structure of the PCNA unloader Elg1-RFC.
Sci Adv, 10, 2024
|
|
8THC
 
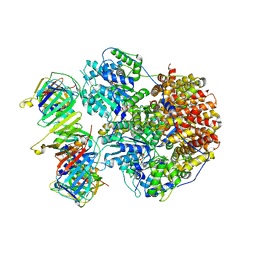 | | Structure of the Saccharomyces cerevisiae clamp unloader Elg1-RFC bound to a cracked PCNA | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ELG1 isoform 1, MAGNESIUM ION, ... | | 著者 | Zheng, F, Yao, Y.N, Georgescu, R, O'Donnell, M.E, Li, H. | | 登録日 | 2023-07-14 | | 公開日 | 2024-05-22 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.67 Å) | | 主引用文献 | Structure of the PCNA unloader Elg1-RFC.
Sci Adv, 10, 2024
|
|
6PTO
 
 | | Structure of Ctf4 trimer in complex with three CMG helicases | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 45, DNA polymerase alpha-binding protein, ... | | 著者 | Yuan, Z, Georgescu, R, Bai, L, Santos, R, Donnell, M, Li, H. | | 登録日 | 2019-07-16 | | 公開日 | 2019-11-20 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (7 Å) | | 主引用文献 | Ctf4 organizes sister replisomes and Pol alpha into a replication factory.
Elife, 8, 2019
|
|
6PTN
 
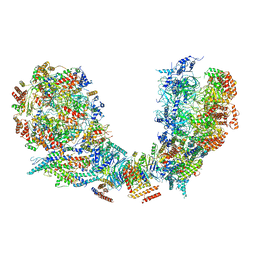 | | Structure of Ctf4 trimer in complex with two CMG helicases | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 45, DNA polymerase alpha-binding protein, ... | | 著者 | Yuan, Z, Georgescu, R, Bai, L, Santos, R, Donnell, M, Li, H. | | 登録日 | 2019-07-16 | | 公開日 | 2019-11-20 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (5.8 Å) | | 主引用文献 | Ctf4 organizes sister replisomes and Pol alpha into a replication factory.
Elife, 8, 2019
|
|
5U8T
 
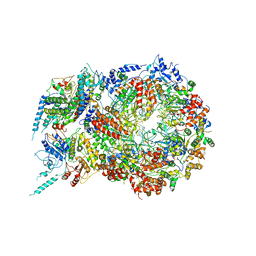 | | Structure of Eukaryotic CMG Helicase at a Replication Fork and Implications | | 分子名称: | Cell division control protein 45, DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DNA replication complex GINS protein PSF1, ... | | 著者 | Li, B, Georgescu, R, Yuan, Z, Santos, R, Sun, J, Zhang, D, Yurieva, O, Li, H, O'Donnell, M.E. | | 登録日 | 2016-12-15 | | 公開日 | 2017-02-08 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (4.9 Å) | | 主引用文献 | Structure of eukaryotic CMG helicase at a replication fork and implications to replisome architecture and origin initiation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5U8S
 
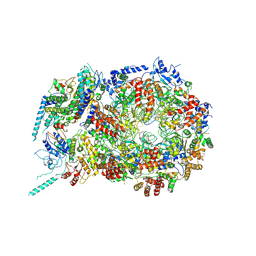 | | Structure of eukaryotic CMG helicase at a replication fork | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 45, DNA (26-MER), ... | | 著者 | Li, H, Li, B, Georgescu, R, Yuan, Z, Santos, R, Sun, J, Zhang, D, Yurieva, O, O'Donnell, M.E. | | 登録日 | 2016-12-14 | | 公開日 | 2017-01-25 | | 最終更新日 | 2020-01-01 | | 実験手法 | ELECTRON MICROSCOPY (6.101 Å) | | 主引用文献 | Structure of eukaryotic CMG helicase at a replication fork and implications to replisome architecture and origin initiation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6PTJ
 
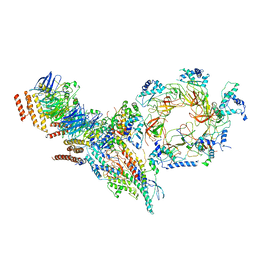 | | Structure of Ctf4 trimer in complex with one CMG helicase | | 分子名称: | Cell division control protein 45, DNA polymerase alpha-binding protein, DNA replication complex GINS protein PSF1, ... | | 著者 | Yuan, Z, Georgescu, R, Bai, L, Santos, R, Donnell, M, Li, H. | | 登録日 | 2019-07-15 | | 公開日 | 2019-11-20 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Ctf4 organizes sister replisomes and Pol alpha into a replication factory.
Elife, 8, 2019
|
|
3JC6
 
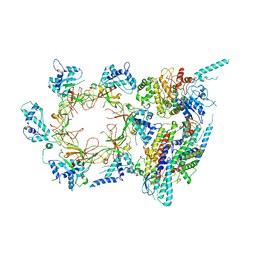 | | Structure of the eukaryotic replicative CMG helicase and pumpjack motion | | 分子名称: | Cell division control protein 45, DNA replication complex GINS protein PSF1, DNA replication complex GINS protein PSF2, ... | | 著者 | Li, H, Bai, L, Yuan, Z, Sun, J, Georgescu, R.E, Liu, J, O'Donnell, M.E. | | 登録日 | 2015-11-24 | | 公開日 | 2016-02-10 | | 最終更新日 | 2018-07-18 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structure of the eukaryotic replicative CMG helicase suggests a pumpjack motion for translocation.
Nat.Struct.Mol.Biol., 23, 2016
|
|
3JC5
 
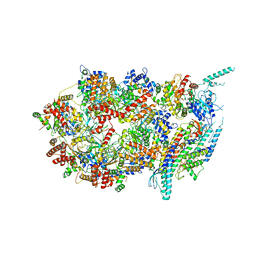 | | Structure of the eukaryotic replicative CMG helicase and pumpjack motion | | 分子名称: | Cell division control protein 45, DNA replication complex GINS protein PSF1, DNA replication complex GINS protein PSF2, ... | | 著者 | Li, H, Bai, L, Yuan, Z, Sun, J, Georgescu, R.E, Liu, J, O'Donnell, M.E. | | 登録日 | 2015-11-24 | | 公開日 | 2016-02-10 | | 最終更新日 | 2018-07-18 | | 実験手法 | ELECTRON MICROSCOPY (4.7 Å) | | 主引用文献 | Structure of the eukaryotic replicative CMG helicase suggests a pumpjack motion for translocation.
Nat.Struct.Mol.Biol., 23, 2016
|
|
3JC7
 
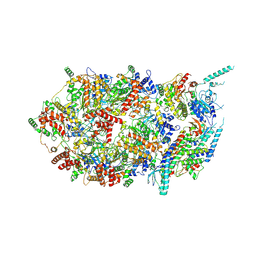 | | Structure of the eukaryotic replicative CMG helicase and pumpjack motion | | 分子名称: | Cell division control protein 45, DNA replication complex GINS protein PSF1, DNA replication complex GINS protein PSF2, ... | | 著者 | Li, H, Bai, L, Yuan, Z, Sun, J, Georgescu, R.E, Liu, J, O'Donnell, M.E. | | 登録日 | 2015-11-24 | | 公開日 | 2016-02-10 | | 最終更新日 | 2018-07-18 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | Structure of the eukaryotic replicative CMG helicase suggests a pumpjack motion for translocation.
Nat.Struct.Mol.Biol., 23, 2016
|
|
2HNH
 
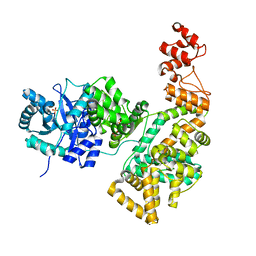 | | Crystal structure of the catalytic alpha subunit of E. coli replicative DNA polymerase III | | 分子名称: | DNA polymerase III alpha subunit, PHOSPHATE ION | | 著者 | Meindert, M.H, Georgescu, R.E, Lee, S, O'Donnell, M, Kuriyan, J. | | 登録日 | 2006-07-12 | | 公開日 | 2006-09-19 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal Structure of the Catalytic alpha Subunit of E. coli Replicative DNA Polymerase III.
Cell(Cambridge,Mass.), 126, 2006
|
|
2HQA
 
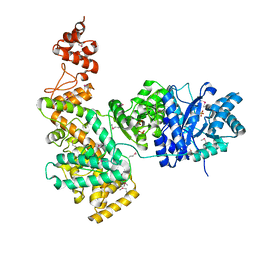 | | Crystal structure of the catalytic alpha subunit of E. Coli replicative DNA polymerase III | | 分子名称: | DNA polymerase III alpha subunit, PHOSPHATE ION | | 著者 | Lamers, M.H, Georgescu, R.E, Lee, S.G, O'Donnell, M, Kuriyan, J. | | 登録日 | 2006-07-18 | | 公開日 | 2006-09-19 | | 最終更新日 | 2017-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal Structure of the Catalytic alpha Subunit of E. coli Replicative DNA Polymerase III.
Cell(Cambridge,Mass.), 126, 2006
|
|
7TFI
 
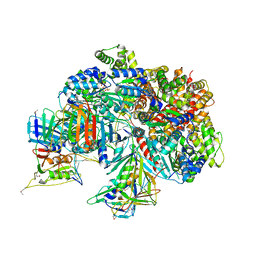 | | Atomic model of the S. cerevisiae clamp-clamp loader complex PCNA-RFC bound to DNA with an open clamp | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | 著者 | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | 登録日 | 2022-01-06 | | 公開日 | 2022-11-16 | | 実験手法 | ELECTRON MICROSCOPY (3.41 Å) | | 主引用文献 | Cryo-EM structures reveal that RFC recognizes both the 3'- and 5'-DNA ends to load PCNA onto gaps for DNA repair.
Elife, 11, 2022
|
|
7TFH
 
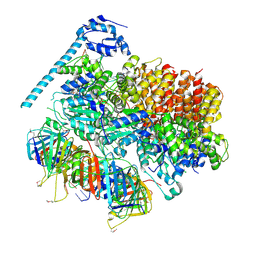 | | Atomic model of the S. cerevisiae clamp-clamp loader complex PCNA-RFC bound to two DNA molecules, one at the 5'-recessed end and the other at the 3'-recessed end | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | 著者 | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | 登録日 | 2022-01-06 | | 公開日 | 2022-11-16 | | 実験手法 | ELECTRON MICROSCOPY (3.09 Å) | | 主引用文献 | Cryo-EM structures reveal that RFC recognizes both the 3'- and 5'-DNA ends to load PCNA onto gaps for DNA repair.
Elife, 11, 2022
|
|
7TFJ
 
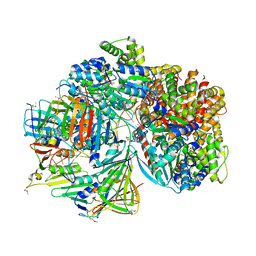 | | Atomic model of S. cerevisiae clamp-clamp loader complex PCNA-RFC bound to DNA with a closed clamp ring | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | 著者 | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | 登録日 | 2022-01-06 | | 公開日 | 2022-11-16 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Cryo-EM structures reveal that RFC recognizes both the 3'- and 5'-DNA ends to load PCNA onto gaps for DNA repair.
Elife, 11, 2022
|
|
