1YHD
 
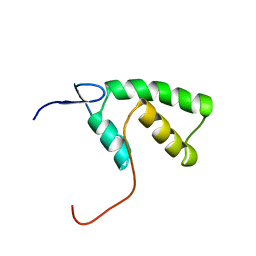 | | The solution structure of YGGX from Escherichia Coli | | Descriptor: | UPF0269 protein yggX | | Authors: | Osborne, M.J, Siddiqui, N, Landgraf, D, Pomposiello, P.J, Gehring, K. | | Deposit date: | 2005-01-07 | | Release date: | 2005-01-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the oxidative stress-related protein YggX from Escherichia coli.
Protein Sci., 14, 2005
|
|
1Y7X
 
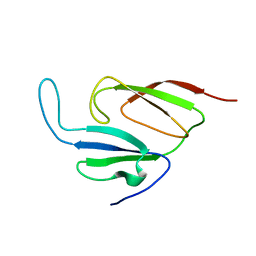 | | Solution structure of a two-repeat fragment of major vault protein | | Descriptor: | Major vault protein | | Authors: | Kozlov, G, Vavelyuk, O, Minailiuc, O, Banville, D, Gehring, K, Ekiel, I. | | Deposit date: | 2004-12-10 | | Release date: | 2005-12-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a two-repeat fragment of major vault protein.
J.Mol.Biol., 356, 2006
|
|
2H8L
 
 | |
2I3E
 
 | |
2ILX
 
 | |
5VRQ
 
 | | Crystal structure of Legionella pneumophila effector AnkC | | Descriptor: | Ankyrin repeat-containing protein | | Authors: | Kozlov, G, Wong, K, Wang, W, Skubak, P, Munoz-Escobar, J, Liu, Y, Pannu, N.S, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2017-05-11 | | Release date: | 2017-11-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.205 Å) | | Cite: | Ankyrin repeats as a dimerization module.
Biochem. Biophys. Res. Commun., 495, 2018
|
|
5WD8
 
 | |
5WD9
 
 | |
3GZH
 
 | |
3PKN
 
 | | Crystal structure of MLLE domain of poly(A) binding protein in complex with PAM2 motif of La-related protein 4 (LARP4) | | Descriptor: | IODIDE ION, La-related protein 4, Polyadenylate-binding protein 1, ... | | Authors: | Xie, J, Kozlov, G, Gehring, K. | | Deposit date: | 2010-11-11 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | La-Related Protein 4 Binds Poly(A), Interacts with the Poly(A)-Binding Protein MLLE Domain via a Variant PAM2w Motif, and Can Promote mRNA Stability.
Mol.Cell.Biol., 31, 2011
|
|
3PT3
 
 | | Crystal structure of the C-terminal lobe of the human UBR5 HECT domain | | Descriptor: | E3 ubiquitin-protein ligase UBR5 | | Authors: | Matta-Camacho, E, Kozlov, G, Menade, M, Gehring, K. | | Deposit date: | 2010-12-02 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure of the HECT C-lobe of the UBR5 E3 ubiquitin ligase.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3IDV
 
 | | Crystal structure of the a0a fragment of ERp72 | | Descriptor: | CHLORIDE ION, Protein disulfide-isomerase A4, ZINC ION | | Authors: | Kozlov, G, Gehring, K. | | Deposit date: | 2009-07-21 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the Catalytic a(0)a Fragment of the Protein Disulfide Isomerase ERp72.
J.Mol.Biol., 401, 2010
|
|
1JH4
 
 | | Solution structure of the C-terminal PABC domain of human poly(A)-binding protein in complex with the peptide from Paip1 | | Descriptor: | polyadenylate-binding protein 1, polyadenylate-binding protein-interacting protein-1 | | Authors: | Kozlov, G, Siddiqui, N, Coillet-Matillon, S, Ekiel, I, Gehring, K. | | Deposit date: | 2001-06-27 | | Release date: | 2003-06-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis of ligand recognition by PABC, a highly specific peptide-binding domain found in poly(A)-binding protein and a HECT ubiquitin ligase
EMBO J., 23, 2004
|
|
3ICH
 
 | |
3ICI
 
 | |
1JGN
 
 | | Solution structure of the C-terminal PABC domain of human poly(A)-binding protein in complex with the peptide from Paip2 | | Descriptor: | polyadenylate-binding protein 1, polyadenylate-binding protein-interacting protein 2 | | Authors: | Kozlov, G, Siddiqui, N, Coillet-Matillon, S, Ekiel, I, Gehring, K. | | Deposit date: | 2001-06-26 | | Release date: | 2003-06-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis of ligand recognition by PABC, a highly specific peptide-binding domain found in poly(A)-binding protein and a HECT ubiquitin ligase
EMBO J., 23, 2004
|
|
3RG0
 
 | | Structural and functional relationships between the lectin and arm domains of calreticulin | | Descriptor: | CALCIUM ION, Calreticulin | | Authors: | Kozlov, G, Pocanschi, C.L, Brockmeier, U, Williams, D.B, Gehring, K. | | Deposit date: | 2011-04-07 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural and Functional Relationships between the Lectin and Arm Domains of Calreticulin.
J.Biol.Chem., 286, 2011
|
|
3PDZ
 
 | |
5V8Z
 
 | |
5V90
 
 | |
4GWR
 
 | |
4F25
 
 | |
4I6X
 
 | |
4IOT
 
 | | High-resolution Structure of Triosephosphate isomerase from E. coli | | Descriptor: | SULFATE ION, Triosephosphate isomerase | | Authors: | Vinaik, R, Kozlov, G, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2013-01-08 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Triosephosphate isomerase is a common crystallization contaminant of soluble His-tagged proteins produced in Escherichia coli.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4F26
 
 | |
