9AUL
 
 | | Structure of SARS-CoV-2 Mpro mutant (A173V,T304I)) in complex with Nirmatrelvir (PF-07321332) | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Gajiwala, K.S, Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2024-02-29 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.421 Å) | | Cite: | In vitro selection and analysis of SARS-CoV-2 nirmatrelvir resistance mutations contributing to clinical virus resistance surveillance.
Sci Adv, 10, 2024
|
|
1DJ8
 
 | |
1DP7
 
 | | COCRYSTAL STRUCTURE OF RFX-DBD IN COMPLEX WITH ITS COGNATE X-BOX BINDING SITE | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DNA (5'-D(*CP*GP*(BRU)P*TP*AP*CP*CP*AP*(BRU)P*GP*GP*TP*AP*AP*CP*G)-3'), ... | | Authors: | Gajiwala, K.S, Chen, H, Cornille, F, Roques, B.P, Reith, W, Mach, B, Burley, S.K. | | Deposit date: | 1999-12-23 | | Release date: | 2000-03-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the winged-helix protein hRFX1 reveals a new mode of DNA binding.
Nature, 403, 2000
|
|
4HZS
 
 | |
4HZR
 
 | | Crystal structure of Ack1 kinase domain | | Descriptor: | 1,2-ETHANEDIOL, Activated CDC42 kinase 1, CHLORIDE ION, ... | | Authors: | Gajiwala, K.S. | | Deposit date: | 2012-11-15 | | Release date: | 2013-02-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Ack1: activation and regulation by allostery.
Plos One, 8, 2013
|
|
5U6B
 
 | | Structure of the Axl kinase domain in complex with a macrocyclic inhibitor | | Descriptor: | (10R)-7-amino-11-chloro-12-fluoro-1-(2-hydroxyethyl)-3,10,16-trimethyl-16,17-dihydro-1H-8,4-(azeno)pyrazolo[4,3-h][2,5,11]benzoxadiazacyclotetradecin-15(10H)-one, Tyrosine-protein kinase receptor UFO | | Authors: | Gajiwala, K.S, Grodsky, N. | | Deposit date: | 2016-12-07 | | Release date: | 2017-07-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | The Axl kinase domain in complex with a macrocyclic inhibitor offers first structural insights into an active TAM receptor kinase.
J. Biol. Chem., 292, 2017
|
|
5U8L
 
 | | Crystal structure of EGFR kinase domain in complex with a sulfonyl fluoride probe XO44 | | Descriptor: | 4-[(4-{4-[(3-cyclopropyl-1H-pyrazol-5-yl)amino]-6-[(prop-2-yn-1-yl)carbamoyl]pyrimidin-2-yl}piperazin-1-yl)methyl]benzene-1-sulfonyl fluoride, Epidermal growth factor receptor, GLYCEROL, ... | | Authors: | Gajiwala, K.S, Ferre, R. | | Deposit date: | 2016-12-14 | | Release date: | 2017-01-25 | | Last modified: | 2017-02-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Broad-Spectrum Kinase Profiling in Live Cells with Lysine-Targeted Sulfonyl Fluoride Probes.
J. Am. Chem. Soc., 139, 2017
|
|
5U6C
 
 | | Crystal structure of the Mer kinase domain in complex with a macrocyclic inhibitor | | Descriptor: | (10R)-7-amino-11-chloro-12-fluoro-1-(2-hydroxyethyl)-3,10,16-trimethyl-16,17-dihydro-1H-8,4-(azeno)pyrazolo[4,3-h][2,5,11]benzoxadiazacyclotetradecin-15(10H)-one, Tyrosine-protein kinase Mer | | Authors: | Gajiwala, K.S, Ferre, R.A. | | Deposit date: | 2016-12-07 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Axl kinase domain in complex with a macrocyclic inhibitor offers first structural insights into an active TAM receptor kinase.
J. Biol. Chem., 292, 2017
|
|
5UG9
 
 | | Crystal structure of the EGFR kinase domain (L858R, T790M, V948R) in complex with a covalent inhibitor N-[(3R,4R)-4-fluoro-1-{6-[(3-methoxy-1-methyl-1H-pyrazol-4-yl)amino]-9-(propan-2-yl)-9H-purin-2-yl}pyrrolidin-3-yl]propanamide | | Descriptor: | 1,2-ETHANEDIOL, Epidermal growth factor receptor, GLYCEROL, ... | | Authors: | Gajiwala, K.S, Ferre, R.A. | | Deposit date: | 2017-01-07 | | Release date: | 2017-03-22 | | Last modified: | 2018-10-10 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Discovery of N-((3R,4R)-4-Fluoro-1-(6-((3-methoxy-1-methyl-1H-pyrazol-4-yl)amino)-9-methyl-9H-purin-2-yl)pyrrolidine-3-yl)acrylamide (PF-06747775) through Structure-Based Drug Design: A High Affinity Irreversible Inhibitor Targeting Oncogenic EGFR Mutants with Selectivity over Wild-Type EGFR.
J. Med. Chem., 60, 2017
|
|
5UGC
 
 | | Crystal structure of the EGFR kinase domain (L858R, T790M, V948R) in complex with a covalent inhibitor N-[(3R,4R)-4-fluoro-1-{6-[(3-methoxy-1-methyl-1H-pyrazol-4-yl)amino]-9-methyl-9H-purin-2-yl}pyrrolidin-3-yl]propanamide | | Descriptor: | 1,2-ETHANEDIOL, Epidermal growth factor receptor, GLYCEROL, ... | | Authors: | Gajiwala, K.S, Ferre, R.A. | | Deposit date: | 2017-01-08 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Discovery of N-((3R,4R)-4-Fluoro-1-(6-((3-methoxy-1-methyl-1H-pyrazol-4-yl)amino)-9-methyl-9H-purin-2-yl)pyrrolidine-3-yl)acrylamide (PF-06747775) through Structure-Based Drug Design: A High Affinity Irreversible Inhibitor Targeting Oncogenic EGFR Mutants with Selectivity over Wild-Type EGFR.
J. Med. Chem., 60, 2017
|
|
5UG8
 
 | | Crystal structure of the EGFR kinase domain (L858R, T790M, V948R) in complex with a covalent inhibitor N-[(3R,4R)-4-fluoro-1-{6-[(1-methyl-1H-pyrazol-4-yl)amino]-9-(propan-2-yl)-9H-purin-2-yl}pyrrolidin-3-yl]propanamide | | Descriptor: | Epidermal growth factor receptor, GLYCEROL, N-[(3R,4R)-4-fluoro-1-{6-[(1-methyl-1H-pyrazol-4-yl)amino]-9-(propan-2-yl)-9H-purin-2-yl}pyrrolidin-3-yl]propanamide, ... | | Authors: | Gajiwala, K.S, Ferre, R.A. | | Deposit date: | 2017-01-07 | | Release date: | 2017-03-22 | | Last modified: | 2017-04-26 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Discovery of N-((3R,4R)-4-Fluoro-1-(6-((3-methoxy-1-methyl-1H-pyrazol-4-yl)amino)-9-methyl-9H-purin-2-yl)pyrrolidine-3-yl)acrylamide (PF-06747775) through Structure-Based Drug Design: A High Affinity Irreversible Inhibitor Targeting Oncogenic EGFR Mutants with Selectivity over Wild-Type EGFR.
J. Med. Chem., 60, 2017
|
|
5UWD
 
 | |
5UGA
 
 | | Crystal structure of the EGFR kinase domain (L858R, T790M, V948R) in complex with 4-(4-{[2-{[(3S)-1-acetylpyrrolidin-3-yl]amino}-9-(propan-2-yl)-9H-purin-6-yl]amino}phenyl)-1-methylpiperazin-1-ium | | Descriptor: | 4-(4-{[2-{[(3S)-1-acetylpyrrolidin-3-yl]amino}-9-(propan-2-yl)-9H-purin-6-yl]amino}phenyl)-1-methylpiperazin-1-ium, Epidermal growth factor receptor, GLYCEROL, ... | | Authors: | Gajiwala, K.S, Ferre, R.A. | | Deposit date: | 2017-01-07 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Discovery of N-((3R,4R)-4-Fluoro-1-(6-((3-methoxy-1-methyl-1H-pyrazol-4-yl)amino)-9-methyl-9H-purin-2-yl)pyrrolidine-3-yl)acrylamide (PF-06747775) through Structure-Based Drug Design: A High Affinity Irreversible Inhibitor Targeting Oncogenic EGFR Mutants with Selectivity over Wild-Type EGFR.
J. Med. Chem., 60, 2017
|
|
5UGB
 
 | |
3K98
 
 | | HSP90 N-terminal domain in complex with (1R)-2-(5-chloro-2,4-dihydroxybenzoyl)-N-ethylisoindoline-1-carboxamide | | Descriptor: | (1R)-2-[(5-chloro-2,4-dihydroxyphenyl)carbonyl]-N-ethyl-2,3-dihydro-1H-isoindole-1-carboxamide, Heat shock protein HSP 90-alpha, PHOSPHATE ION | | Authors: | Gajiwala, K.S, Davies II, J.F. | | Deposit date: | 2009-10-15 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dihydroxyphenylisoindoline amides as orally bioavailable inhibitors of the heat shock protein 90 (hsp90) molecular chaperone.
J.Med.Chem., 53, 2010
|
|
3K99
 
 | | HSP90 N-terminal domain in complex with 4-(1,3-dihydro-2H-isoindol-2-ylcarbonyl)benzene-1,3-diol | | Descriptor: | 4-(1,3-dihydro-2H-isoindol-2-ylcarbonyl)benzene-1,3-diol, Heat shock protein HSP 90-alpha | | Authors: | Gajiwala, K.S, Davies II, J.F. | | Deposit date: | 2009-10-15 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dihydroxyphenylisoindoline amides as orally bioavailable inhibitors of the heat shock protein 90 (hsp90) molecular chaperone.
J.Med.Chem., 53, 2010
|
|
3K97
 
 | |
3HEK
 
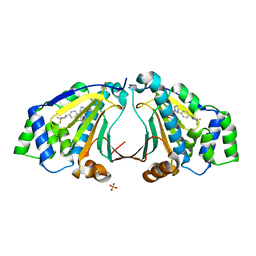 | |
3EKR
 
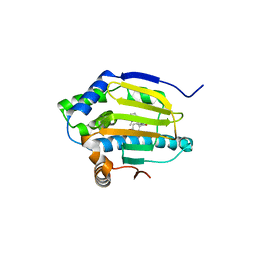 | | Dihydroxylphenyl amides as inhibitors of the Hsp90 molecular chaperone | | Descriptor: | 4-{[(2R)-2-(2-methylphenyl)pyrrolidin-1-yl]carbonyl}benzene-1,3-diol, Heat shock protein HSP 90-alpha, PHOSPHATE ION | | Authors: | Gajiwala, K.S. | | Deposit date: | 2008-09-19 | | Release date: | 2008-11-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dihydroxylphenyl amides as inhibitors of the Hsp90 molecular chaperone.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3EKO
 
 | |
1TAE
 
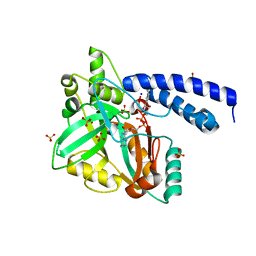 | |
1TA8
 
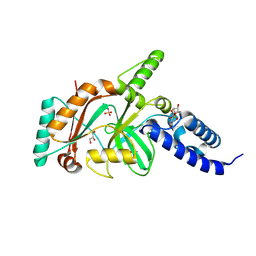 | |
1MDO
 
 | | Crystal structure of ArnB aminotransferase with pyridomine 5' phosphate | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ArnB aminotransferase | | Authors: | Noland, B.W, Newman, J.M, Hendle, J, Badger, J, Christopher, J.A, Tresser, J, Buchanan, M.D, Wright, T, Rutter, M.E, Sanderson, W.E, Muller-Dieckmann, H.-J, Gajiwala, K, Sauder, J.M, Buchanan, S.G. | | Deposit date: | 2002-08-07 | | Release date: | 2002-12-11 | | Last modified: | 2018-12-26 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural studies of Salmonella typhimurium ArnB (PmrH) aminotransferase: A 4-amino-4-deoxy-L-arabinose lipopolysaccharide modifying enzyme
Structure, 10, 2002
|
|
3RLQ
 
 | | Co-crystal structure of the HSP90 ATP binding domain in complex with 4-(2,4-dichloro-5-methoxyphenyl)-2-methyl-7H-pyrrolo[2,3-d]pyrimidine-5- carbonitrile | | Descriptor: | 4-(2,4-dichloro-5-methoxyphenyl)-2-methyl-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, Heat shock protein HSP 90-alpha, PHOSPHATE ION | | Authors: | Kung, P.-P, Sinnema, P.-J, Richardson, P, Hickey, M.J, Gajiwala, K.S, Wang, F, Huang, B, McClellan, G, Wang, J, Maegley, K, Bergqvist, S, Mehta, P.P, Kania, R. | | Deposit date: | 2011-04-20 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design strategies to target crystallographic waters applied to the Hsp90 molecular chaperone.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3RLP
 
 | | Co-crystal structure of the HSP90 ATP binding domain in complex with 4-(2,4-dichloro-5-methoxyphenyl)-6-methylpyrimidin-2-amine | | Descriptor: | 4-(2,4-dichloro-5-methoxyphenyl)-6-methylpyrimidin-2-amine, Heat shock protein HSP 90-alpha, PHOSPHATE ION | | Authors: | Kung, P.-P, Sinnema, P.-J, Richardson, P, Hickey, M.J, Gajiwala, K.S, Wang, F, Huang, B, McClellan, G, Wang, J, Maegley, K, Bergqvist, S, Mehta, P.P, Kania, R. | | Deposit date: | 2011-04-20 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design strategies to target crystallographic waters applied to the Hsp90 molecular chaperone.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
