7C9N
 
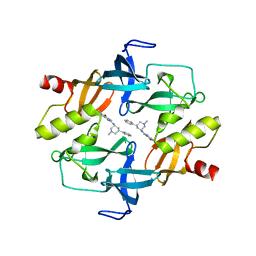 | | Crystal structure of SETDB1 tudor domain in complexed with Compound 1. | | Descriptor: | 3,5-dimethyl-2-[[(3R,5R)-1-methyl-5-phenyl-piperidin-3-yl]amino]pyrrolo[3,2-d]pyrimidin-4-one, Histone-lysine N-methyltransferase SETDB1 | | Authors: | Guo, Y, Xiong, L, Mao, X, Yang, S. | | Deposit date: | 2020-06-06 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.472 Å) | | Cite: | Structure-Guided Discovery of a Potent and Selective Cell-Active Inhibitor of SETDB1 Tudor Domain.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7XGC
 
 | |
7XGQ
 
 | |
7CJU
 
 | |
3NFP
 
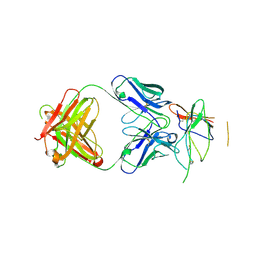 | | Crystal structure of the Fab fragment of therapeutic antibody daclizumab in complex with IL-2Ra (CD25) ectodomain | | Descriptor: | Heavy chain of Fab fragment of daclizumab, Interleukin-2 receptor subunit alpha, Light chain of Fab fragment of daclizumab | | Authors: | Yang, H, Wang, J, Du, J, Zhong, C, Guo, Y, Ding, J. | | Deposit date: | 2010-06-10 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structural basis of immunosuppression by the therapeutic antibody daclizumab
Cell Res., 20, 2010
|
|
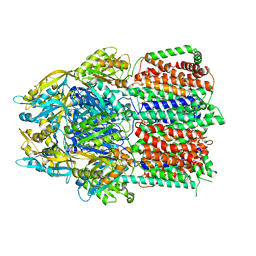
6CSX
 
 | |
4LZ6
 
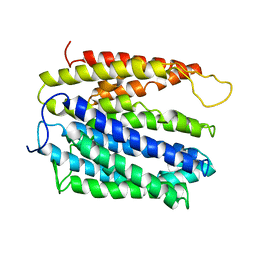 | | Structure of MATE multidrug transporter DinF-BH | | Descriptor: | BH2163 protein | | Authors: | Lu, M, Radchenko, M, Symersky, J, Nie, R, Guo, Y. | | Deposit date: | 2013-07-31 | | Release date: | 2013-10-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into H(+)-coupled multidrug extrusion by a MATE transporter
Nat.Struct.Mol.Biol., 20, 2013
|
|
4LZ9
 
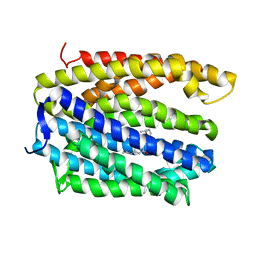 | | Structure of MATE multidrug transporter DinF-BH in complex with R6G | | Descriptor: | BH2163 protein, RHODAMINE 6G | | Authors: | Lu, M, Radchenko, M, Symersky, J, Nie, R, Guo, Y. | | Deposit date: | 2013-07-31 | | Release date: | 2013-10-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural insights into H(+)-coupled multidrug extrusion by a MATE transporter
Nat.Struct.Mol.Biol., 20, 2013
|
|
5VKQ
 
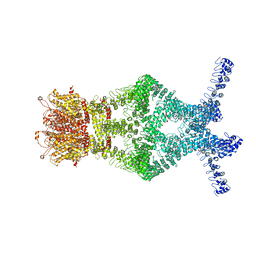 | | Structure of a mechanotransduction ion channel Drosophila NOMPC in nanodisc | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, No mechanoreceptor potential C isoform L | | Authors: | Jin, P, Bulkley, D, Guo, Y, Zhang, W, Guo, Z, Huynh, W, Wu, S, Meltzer, S, Chen, T, Jan, L.Y, Jan, Y.-N, Cheng, Y. | | Deposit date: | 2017-04-22 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Electron cryo-microscopy structure of the mechanotransduction channel NOMPC.
Nature, 547, 2017
|
|
8HLD
 
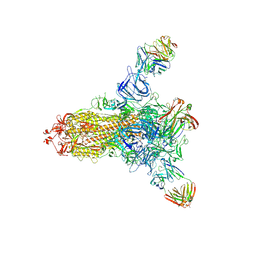 | | S protein of SARS-CoV-2 in complex with 26434 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Zhang, Y.Y, Guo, Y.Y, Zhou, Q. | | Deposit date: | 2022-11-29 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | A SARS-CoV-2 Spike N-terminal domain neutralizing antibody targets on a glycans shielded silent face and inhibits virus entry via hindering the recognition of RBD and hACE2
To Be Published
|
|
8HLC
 
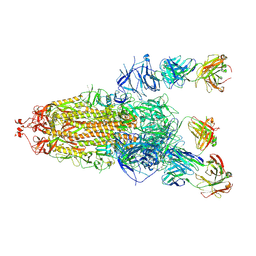 | | S protein of SARS-CoV-2 in complex with 3711 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, Y.Y, Guo, Y.Y, Zhou, Q. | | Deposit date: | 2022-11-29 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | A SARS-CoV-2 Spike N-terminal domain neutralizing antibody targets on a glycans shielded silent face and inhibits virus entry via hindering the recognition of RBD and hACE2
To Be Published
|
|
5EN0
 
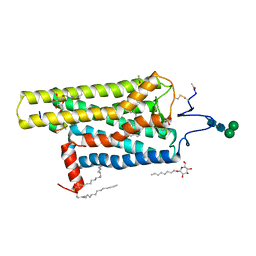 | | Crystal Structure of T94I rhodopsin mutant | | Descriptor: | ACETATE ION, Guanine nucleotide-binding protein G(t) subunit alpha-3, PALMITIC ACID, ... | | Authors: | Singhal, A, Guo, Y, Matkovic, M, Schertler, G, Deupi, X, Yan, E, Standfuss, J. | | Deposit date: | 2015-11-08 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural role of the T94I rhodopsin mutation in congenital stationary night blindness.
Embo Rep., 17, 2016
|
|
8Z5L
 
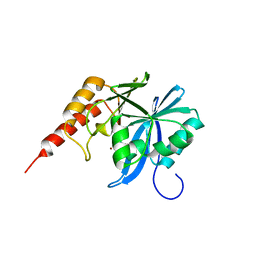 | | Crystal structure of metallo-beta-lactamse, IMP-1, complexed with a quinolinone-based inhibitor | | Descriptor: | 3-[2-azanyl-5-[2-cyclohexylethyl-[3-(4-methylphenoxy)propyl]amino]phenyl]propanoic acid, Metallo-beta-lactamase type 2, ZINC ION | | Authors: | Kamo, T, Kuroda, K, Nimura, S, Guo, Y, Kondo, S, Nukaga, M, Hoshino, T. | | Deposit date: | 2024-04-18 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Development of Inhibitory Compounds for Metallo-beta-lactamase through Computational Design and Crystallographic Analysis.
Biochemistry, 63, 2024
|
|
7BTK
 
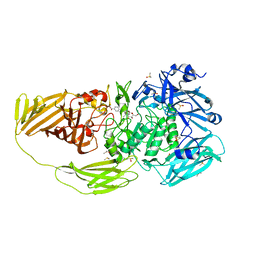 | | E.coli beta-galactosidase (E537Q) in complex with fluorescent probe KSA01 | | Descriptor: | 4-[[2-[(E)-2-[4-[(2S,3R,4S,5R,6R)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxyphenyl]ethenyl]-3,3-dimethyl-2H-indol-1-yl]methyl]benzoic acid, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Chen, X, Hu, Y.L, Liu, Q.M, Gao, Y, Yuan, R, Guo, Y. | | Deposit date: | 2020-04-01 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Two-Dimensional Design Strategy to Construct Smart Fluorescent Probes for the Precise Tracking of Senescence.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7BRS
 
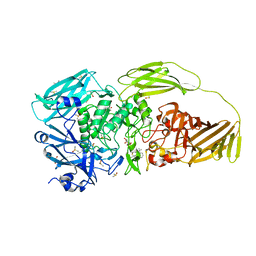 | | E.coli beta-galactosidase (E537Q) in complex with fluorescent probe KSA02 | | Descriptor: | 8-[2-[(E)-2-[4-[(2S,3R,4S,5R,6R)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxyphenyl]ethenyl]-3,3-dimethyl-indol-1-ium-1-yl]octanoic acid, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Chen, X, Hu, Y.L, Gao, Y, Yuan, R, Guo, Y. | | Deposit date: | 2020-03-30 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Two-Dimensional Design Strategy to Construct Smart Fluorescent Probes for the Precise Tracking of Senescence.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7N4T
 
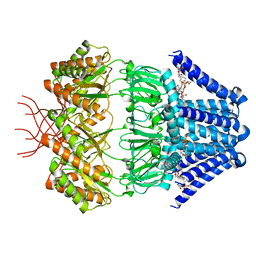 | | Low conductance mechanosensitive channel YnaI | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Low conductance mechanosensitive channel YnaI | | Authors: | Catalano, C, Ben-Hail, D, Qiu, W, des Georges, A, Guo, Y. | | Deposit date: | 2021-06-04 | | Release date: | 2022-04-20 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Cryo-EM Structure of Mechanosensitive Channel YnaI Using SMA2000: Challenges and Opportunities.
Membranes (Basel), 11, 2021
|
|
7BQ9
 
 | | Crystal structure of ASFV p15 | | Descriptor: | 60 kDa polyprotein | | Authors: | Fu, D, Chen, C, Guo, Y. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-24 | | Method: | X-RAY DIFFRACTION (2.612 Å) | | Cite: | Structure of a bifunctional membrane-DNA binding protein, African swine fever virus p15
Protein Cell, 2020
|
|
6VSC
 
 | |
6VSA
 
 | |
7BQA
 
 | | Crystal structure of ASFV p35 | | Descriptor: | 60 kDa polyprotein | | Authors: | Li, G.B, Fu, D, Chen, C, Guo, Y. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-24 | | Last modified: | 2021-05-05 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Crystal structure of the African swine fever virus structural protein p35 reveals its role for core shell assembly.
Protein Cell, 11, 2020
|
|
3GAS
 
 | | Crystal Structure of Helicobacter pylori Heme Oxygenase Hugz in Complex with Heme | | Descriptor: | 1,2-ETHANEDIOL, AZIDE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Jiang, F, Hu, Y.L, Guo, Y, Guo, G, Zou, Q.M, Wang, D.C. | | Deposit date: | 2009-02-18 | | Release date: | 2010-02-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Helicobacter pylori Heme Oxygenase Hugz in Complex with Heme
To be Published
|
|
6VRS
 
 | | Single particle reconstruction of glucose isomerase from Streptomyces rubiginosus based on data acquired in the presence of substantial aberrations | | Descriptor: | MANGANESE (II) ION, xylose isomerase | | Authors: | Bromberg, R, Guo, Y, Borek, D, Otwinowski, Z. | | Deposit date: | 2020-02-09 | | Release date: | 2020-02-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | High-resolution cryo-EM reconstructions in the presence of substantial aberrations
Iucrj, 7, 2020
|
|
5M11
 
 | | Structural and functional probing of PorZ, an essential bacterial surface component of the type-IX secretion system of human oral-microbiomic Porphyromonas gingivalis. | | Descriptor: | CACODYLATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Lasica, A.M, Goulas, T, Mizgalska, D, Zhou, X, Ksiazek, M, Madej, M, Guo, Y, Guevara, T, Nowak, M, Potempa, B, Goel, A, Sztukowska, M, Prabhakar, A.T, Bzowska, M, Widziolek, M, Thogersen, I.B, Enghild, J.J, Simonian, M, Kulczyk, A.W, Nguyen, K.-A, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2016-10-07 | | Release date: | 2016-11-09 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and functional probing of PorZ, an essential bacterial surface component of the type-IX secretion system of human oral-microbiomic Porphyromonas gingivalis.
Sci Rep, 6, 2016
|
|
5VBT
 
 | | Crystal structure of a highly specific and potent USP7 ubiquitin variant inhibitor | | Descriptor: | UBH04 | | Authors: | DONG, A, DONG, X, LIU, L, GUO, Y, LI, Y, ZHANG, W, WALKER, J.R, SIDHU, S, Bountra, C, Arrowsmith, C.H, Edwards, A.M, TONG, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-03-30 | | Release date: | 2017-06-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystal structure of a highly specific and potent USP7 ubiquitin variant inhibitor
to be published
|
|
7RR7
 
 | | Multidrug efflux pump subunit AcrB | | Descriptor: | DODECANE, Multidrug efflux pump subunit AcrB, PHOSPHATIDYLETHANOLAMINE | | Authors: | Trinh, T.K.H, Cabezas, A, Catalano, C, Qiu, W, des Georges, A, Guo, Y. | | Deposit date: | 2021-08-09 | | Release date: | 2022-08-17 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Multidrug efflux pump subunit AcrB
To Be Published
|
|
