4BCB
 
 | | PROLYL OLIGOPEPTIDASE FROM PORCINE BRAIN WITH A COVALENTLY BOUND P2- substituted N-acyl-prolylpyrrolidine inhibitor | | Descriptor: | (5R,6R,8S)-8-(3-{[AMINO(IMINO)METHYL]AMINO}PHENYL)-5-CYCLOHEXYL-6-HYDROXY-3-OXO-1-PHENYL-2,7-DIOXA-4-AZA-6-PHOSPHANONAN-9-OIC ACID 6-OXIDE, GLYCEROL, PROLYL ENDOPEPTIDASE, ... | | Authors: | VanDerVeken, P, Fulop, V, Rea, D, Gerard, M, VanElzen, R, Joossens, J, Cheng, J.D, Baekelandt, V, DeMeester, I, Lambeir, A.M, Augustyns, K. | | Deposit date: | 2012-10-01 | | Release date: | 2013-03-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | P2-Substituted N-Acylprolylpyrrolidine Inhibitors of Prolyl Oligopeptidase: Biochemical Evaluation, Binding Mode Determination, and Assessment in a Cellular Model of Synucleinopathy.
J.Med.Chem., 55, 2012
|
|
4BCC
 
 | | PROLYL OLIGOPEPTIDASE FROM PORCINE BRAIN WITH A COVALENTLY BOUND P2- substituted N-acyl-prolylpyrrolidine inhibitor | | Descriptor: | GLYCEROL, PROLYL ENDOPEPTIDASE, TRIS(HYDROXYETHYL)AMINOMETHANE, ... | | Authors: | VanDerVeken, P, Fulop, V, Rea, D, Gerard, M, VanElzen, R, Joossens, J, Cheng, J.D, Baekelandt, V, DeMeester, I, Lambeir, A.M, Augustyns, K. | | Deposit date: | 2012-10-01 | | Release date: | 2013-03-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | P2-Substituted N-Acylprolylpyrrolidine Inhibitors of Prolyl Oligopeptidase: Biochemical Evaluation, Binding Mode Determination, and Assessment in a Cellular Model of Synucleinopathy.
J.Med.Chem., 55, 2012
|
|
4BT5
 
 | | acetolactate decarboxylase with a bound (2S,3R)-2,3-Dihydroxy-2- methylbutanoic acid | | Descriptor: | (2S,3R)-2,3-dihydroxy-2-methylbutanoic acid, ALPHA-ACETOLACTATE DECARBOXYLASE, ZINC ION | | Authors: | A Marlow, V, Rea, D, Najmudin, S, Wills, M, Fulop, V. | | Deposit date: | 2013-06-12 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure and Mechanism of Acetolactate Decarboxylase.
Acs Chem.Biol., 8, 2013
|
|
4BT7
 
 | | acetolactate decarboxylase with a bound phosphate ion | | Descriptor: | ALPHA-ACETOLACTATE DECARBOXYLASE, PHOSPHATE ION, ZINC ION | | Authors: | A Marlow, V, Rea, D, Najmudin, S, Wills, M, Fulop, V. | | Deposit date: | 2013-06-12 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure and Mechanism of Acetolactate Decarboxylase.
Acs Chem.Biol., 8, 2013
|
|
4BT3
 
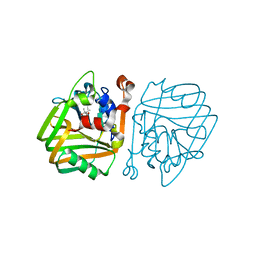 | | acetolactate decarboxylase with a bound (2R,3R)-2,3-Dihydroxy-2- methylbutanoic acid | | Descriptor: | (2R,3R)-2,3-Dihydroxy-2-methylbutanoic acid, ALPHA-ACETOLACTATE DECARBOXYLASE, ZINC ION | | Authors: | A Marlow, V, Rea, D, Najmudin, S, Wills, M, Fulop, V. | | Deposit date: | 2013-06-12 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure and Mechanism of Acetolactate Decarboxylase.
Acs Chem.Biol., 8, 2013
|
|
4BT6
 
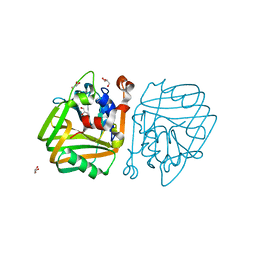 | | acetolactate decarboxylase with a bound glycerol | | Descriptor: | ALPHA-ACETOLACTATE DECARBOXYLASE, GLYCEROL, ZINC ION | | Authors: | A Marlow, V, Rea, D, Najmudin, S, Wills, M, Fulop, V. | | Deposit date: | 2013-06-12 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and Mechanism of Acetolactate Decarboxylase.
Acs Chem.Biol., 8, 2013
|
|
4BT4
 
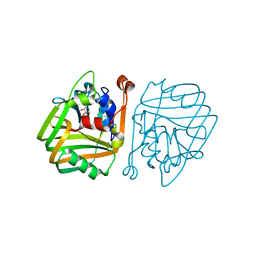 | | acetolactate decarboxylase with a bound (2S,3S)-2,3-Dihydroxy-2- methylbutanoic acid | | Descriptor: | (2S,3S)-2,3-dihydroxy-2-methylbutanoic acid, ALPHA-ACETOLACTATE DECARBOXYLASE, ZINC ION | | Authors: | A Marlow, V, Rea, D, Najmudin, S, Wills, M, Fulop, V. | | Deposit date: | 2013-06-12 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and Mechanism of Acetolactate Decarboxylase.
Acs Chem.Biol., 8, 2013
|
|
4BT2
 
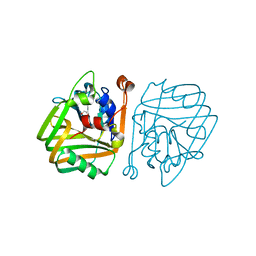 | | acetolactate decarboxylase with a bound 1,2-ETHANEDIOL | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-ACETOLACTATE DECARBOXYLASE, ZINC ION | | Authors: | A Marlow, V, Rea, D, Najmudin, S, Wills, M, Fulop, V. | | Deposit date: | 2013-06-12 | | Release date: | 2013-09-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure and Mechanism of Acetolactate Decarboxylase
Acs Chem.Biol., 8, 2013
|
|
4C5B
 
 | | The X-ray crystal structure of D-alanyl-D-alanine ligase in complex with ATP and D-ala-D-ala | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CARBONATE ION, D-ALANINE, ... | | Authors: | Batson, S, Majce, V, Lloyd, A.J, Rea, D, Fishwick, C.W.G, Simmons, K.J, Fulop, V, Roper, D.I. | | Deposit date: | 2013-09-10 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Inhibition of D-Ala:D-Ala ligase through a phosphorylated form of the antibiotic D-cycloserine.
Nat Commun, 8, 2017
|
|
4C5C
 
 | | The X-ray crystal structure of D-alanyl-D-alanine ligase in complex with ADP and D-ala-D-ala | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, D-ALANINE, D-ALANINE--D-ALANINE LIGASE, ... | | Authors: | Batson, S, Majce, V, Lloyd, A.J, Rea, D, Fishwick, C.W.G, Simmons, K.J, Fulop, V, Roper, D.I. | | Deposit date: | 2013-09-10 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Inhibition of D-Ala:D-Ala ligase through a phosphorylated form of the antibiotic D-cycloserine.
Nat Commun, 8, 2017
|
|
4C5A
 
 | | The X-ray crystal structures of D-alanyl-D-alanine ligase in complex ADP and D-cycloserine phosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, D-ALANINE--D-ALANINE LIGASE, GLYCEROL, ... | | Authors: | Batson, S, Majce, V, Lloyd, A.J, Rea, D, Fishwick, C.W.G, Simmons, K.J, Fulop, V, Roper, D.I. | | Deposit date: | 2013-09-10 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Inhibition of D-Ala:D-Ala ligase through a phosphorylated form of the antibiotic D-cycloserine.
Nat Commun, 8, 2017
|
|
2VC3
 
 | |
2X30
 
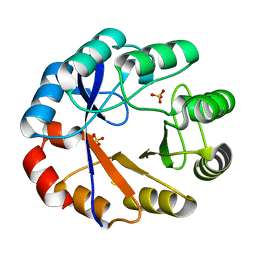 | | Crystal structure of the r139n mutant of a bifunctional enzyme pria | | Descriptor: | PHOSPHORIBOSYL ISOMERASE A, SULFATE ION | | Authors: | Noda-Garcia, L, Camacho-Zarco, A.R, Verdel-Aranda, K, Wright, H, Soberon, X, Fulop, V, Barona-Gomez, F. | | Deposit date: | 2010-01-20 | | Release date: | 2010-02-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification and Analysis of Residues Contained on Beta --> Alpha Loops of the Dual-Substrate (Betaalpha)(8) Phosphoribosyl Isomerase a (Pria) Specific for its Phosphoribosyl Anthranilate Isomerase Activity.
Protein Sci., 19, 2010
|
|
2VEP
 
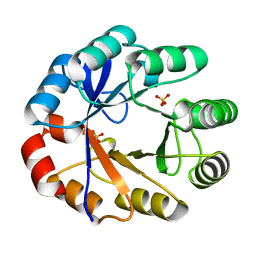 | | Crystal Structure Of The Full Length Bifunctional Enzyme Pria | | Descriptor: | PHOSPHORIBOSYL ISOMERASE A, SULFATE ION | | Authors: | Wright, H, Noda-Garcia, L, Ochoa-Leyva, A, Hodgson, D.A, Fulop, V, Barona-Gomez, F. | | Deposit date: | 2007-10-25 | | Release date: | 2007-11-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure/Function Relationship of a Dual Substrate (Betaalpha)(8)-Isomerase
Biochem.Biophys.Res.Commun., 365, 2008
|
|
2XDW
 
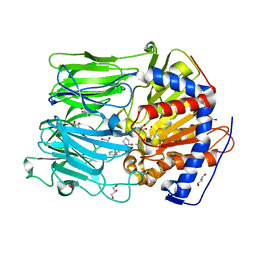 | | Inhibition of Prolyl Oligopeptidase with a Synthetic Unnatural Dipeptide | | Descriptor: | GLYCEROL, PROLYL ENDOPEPTIDASE, SYNTHETIC PEPTIDE PHQ-PRO-YCP, ... | | Authors: | Racys, D.T, Rea, D, Fulop, V, Wills, M. | | Deposit date: | 2010-05-09 | | Release date: | 2010-06-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Inhibition of Prolyl Oligopeptidase with a Synthetic Unnatural Dipeptide
Bioorg.Med.Chem., 18, 2010
|
|
2VHD
 
 | | Crystal Structure Of The Di-Haem Cytochrome C Peroxidase From Pseudomonas aeruginosa - Mixed Valence Form | | Descriptor: | CALCIUM ION, CYTOCHROME C551 PEROXIDASE, HEME C | | Authors: | Echalier, A, Brittain, T, Wright, J, Boycheva, S, Mortuza, G.B, Fulop, V, Watmough, N.J. | | Deposit date: | 2007-11-20 | | Release date: | 2008-02-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Redox-Linked Structural Changes Associated with the Formation of a Catalytically Competent Form of the Diheme Cytochrome C Peroxidase from Pseudomonas Aeruginosa
Biochemistry, 47, 2008
|
|
2VC4
 
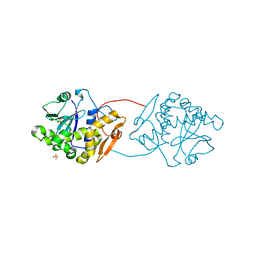 | | Ricin A-Chain (Recombinant) E177D Mutant | | Descriptor: | GLYCEROL, RICIN A CHAIN, SULFATE ION | | Authors: | Marsden, C.J, Fulop, V. | | Deposit date: | 2007-09-18 | | Release date: | 2007-10-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | The Isolation and Characterisation of Temperature-Dependent Ricin a Chain Molecules in Saccharomyces Cerevisiae
FEBS J., 274, 2007
|
|
4AXK
 
 | | CRYSTAL STRUCTURE OF subHisA from the thermophile Corynebacterium efficiens | | Descriptor: | 1-(5-PHOSPHORIBOSYL)-5-((5'-PHOSPHORIBOSYLAMINO) METHYLIDENEAMINO)IMIDAZOLE-4-CARBOXAMIDE ISOMERASE, GLYCEROL | | Authors: | Noda-Garcia, L, Camacho-Zarco, A.R, Medina-Ruiz, S, Verduzco-Castro, E.A, Gaytan, P, Carrillo-Tripp, M, Fulop, V, Barona-Gomez, F. | | Deposit date: | 2012-06-13 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Evolution of Substrate Specificity in a Recipient'S Enzyme Following Horizontal Gene Transfer.
Mol.Biol.Evol., 30, 2013
|
|
1AOQ
 
 | |
4BP8
 
 | |
4AN1
 
 | | PROLYL OLIGOPEPTIDASE FROM PORCINE BRAIN WITH A COVALENTLY BOUND INHIBITOR IC-4 | | Descriptor: | (2S)-N-benzyl-2-({(2S)-2-[(1R)-1,2-dihydroxyethyl]pyrrolidin-1-yl}carbonyl)pyrrolidine-1-carboxamide, GLYCEROL, PROLYL ENDOPEPTIDASE | | Authors: | Kaszuba, K, Rog, T, Danne, R, Canning, P, Fulop, V, Juhasz, T, Szeltner, Z, St-Pierre, J.F, Garcia-Horsman, A, Mannisto, P.T, Karttunen, M, Hokkanen, J, Bunker, A. | | Deposit date: | 2012-03-14 | | Release date: | 2012-05-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Dynamics, Crystallography and Mutagenesis Studies on the Substrate Gating Mechanism of Prolyl Oligopeptidase.
Biochimie, 94, 2012
|
|
4AN0
 
 | | PROLYL OLIGOPEPTIDASE FROM PORCINE BRAIN WITH A COVALENTLY BOUND INHIBITOR IC-3 | | Descriptor: | (2S)-1-[1-(4-phenylbutanoyl)-L-prolyl]pyrrolidine-2-carbonitrile, GLYCEROL, PROLYL ENDOPEPTIDASE | | Authors: | Kaszuba, K, Rog, T, Danne, R, Canning, P, Fulop, V, Juhasz, T, Szeltner, Z, St-Pierre, J.F, Garcia-Horsman, A, Mannisto, P.T, Karttunen, M, Hokkanen, J, Bunker, A. | | Deposit date: | 2012-03-14 | | Release date: | 2012-05-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular dynamics, crystallography and mutagenesis studies on the substrate gating mechanism of prolyl oligopeptidase.
Biochimie, 94, 2012
|
|
4AMY
 
 | | PROLYL OLIGOPEPTIDASE FROM PORCINE BRAIN WITH A COVALENTLY BOUND INHIBITOR IC-1 | | Descriptor: | 1-[(2R,5S)-2-tert-butyl-5-({(2S)-2-[(1R)-1,2-dihydroxyethyl]pyrrolidin-1-yl}carbonyl)pyrrolidin-1-yl]-4-phenylbutan-1-one, GLYCEROL, PROLYL ENDOPEPTIDASE | | Authors: | Kaszuba, K, Rog, T, Danne, R, Canning, P, Fulop, V, Juhasz, T, Szeltner, Z, St-Pierre, J.F, Garcia-Horsman, A, Mannisto, P.T, Karttunen, M, Hokkanen, J, Bunker, A. | | Deposit date: | 2012-03-14 | | Release date: | 2012-05-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Dynamics, Crystallography and Mutagenesis Studies on the Substrate Gating Mechanism of Prolyl Oligopeptidase.
Biochimie, 94, 2012
|
|
4AMZ
 
 | | PROLYL OLIGOPEPTIDASE FROM PORCINE BRAIN WITH A COVALENTLY BOUND INHIBITOR IC-2 | | Descriptor: | (5R)-N-benzyl-5-({(2S)-2-[(1R)-1,2-dihydroxyethyl]pyrrolidin-1-yl}carbonyl)cyclopent-1-ene-1-carboxamide, GLYCEROL, PROLYL ENDOPEPTIDASE | | Authors: | Kaszuba, K, Rog, T, Danne, R, Canning, P, Fulop, V, Juhasz, T, Szeltner, Z, St-Pierre, J.F, Garcia-Horsman, A, Mannisto, P.T, Karttunen, M, Hokkanen, J, Bunker, A. | | Deposit date: | 2012-03-14 | | Release date: | 2012-05-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Dynamics, Crystallography and Mutagenesis Studies on the Substrate Gating Mechanism of Prolyl Oligopeptidase.
Biochimie, 94, 2012
|
|
4AX4
 
 | | PROLYL OLIGOPEPTIDASE FROM PORCINE BRAIN, H680A MUTANT | | Descriptor: | GLYCEROL, PROLYL OLIGOPEPTIDASE | | Authors: | Szeltner, Z, Juhasz, T, Szamosi, I, Rea, D, Fulop, V, Modos, K, Juliano, L, Polgar, L. | | Deposit date: | 2012-06-08 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Loops Facing the Active Site of Prolyl Oligopeptidase are Crucial Components in Substrate Gating and Specificity.
Biochim.Biophys.Acta, 1834, 2012
|
|
