7KJW
 
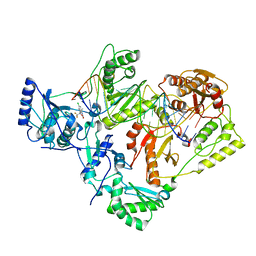 | | Structure of HIV-1 reverse transcriptase initiation complex core with efavirenz | | Descriptor: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, HIV-1 viral RNA fragment, MAGNESIUM ION, ... | | Authors: | Ha, B, Larsen, K.P, Zhang, J, Fu, Z, Montabana, E, Jackson, L.N, Chen, D.H, Puglisi, E.V. | | Deposit date: | 2020-10-26 | | Release date: | 2021-03-17 | | Last modified: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | High-resolution view of HIV-1 reverse transcriptase initiation complexes and inhibition by NNRTI drugs.
Nat Commun, 12, 2021
|
|
2P8T
 
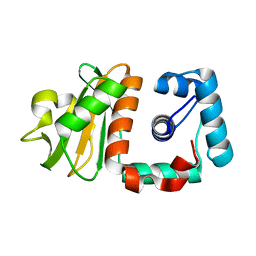 | | Hypothetical protein PH0730 from Pyrococcus horikoshii OT3 | | Descriptor: | Hypothetical protein PH0730 | | Authors: | Chen, L, Zhao, M, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Zhu, J, Swindell, J.T, Fu, Z.-Q, Chrzas, J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-23 | | Release date: | 2007-04-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hypothetical protein PH0730 from Pyrococcus horikoshii OT3
To be Published
|
|
2P17
 
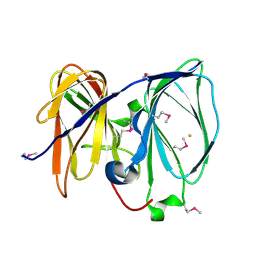 | | Crystal structure of GK1651 from Geobacillus kaustophilus | | Descriptor: | FE (III) ION, Pirin-like protein | | Authors: | Zhu, J, Swindell II, J.T, Chen, L, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Fu, Z.-Q, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-02 | | Release date: | 2007-05-01 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal structure of GK1651 from Geobacillus kaustophilus
To be Published
|
|
8FF2
 
 | |
8FF3
 
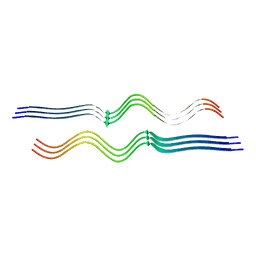 | |
7WFC
 
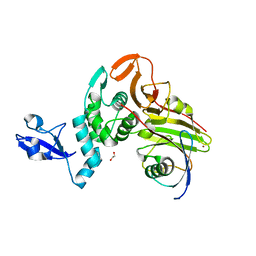 | | X-ray structure of HKU1-PLP2(Cys109Ser) catalytic mutant in complex with free ubiquitin | | Descriptor: | 1,2-ETHANEDIOL, 60S ribosomal protein L40, Papain-like protease, ... | | Authors: | Xiong, Y.X, Fu, Z.Y, Huang, H. | | Deposit date: | 2021-12-26 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The substrate selectivity of papain-like proteases from human-infecting coronaviruses correlates with innate immune suppression.
Sci.Signal., 16, 2023
|
|
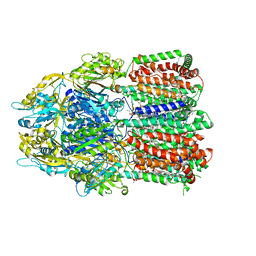
6BAJ
 
 | |
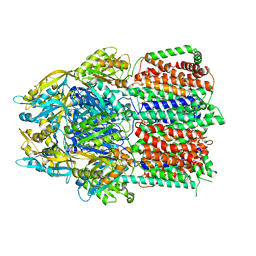
6CSX
 
 | |
8GWS
 
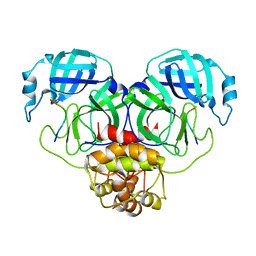 | | SARS-CoV-2 Mpro 1-302 c145a in complex with peptide 4 | | Descriptor: | Replicase polyprotein 1ab, VAL-LYS-LEU-GLN-ALA-ILE-PHE-ARG | | Authors: | Liu, M, Fu, Z, Huang, H. | | Deposit date: | 2022-09-17 | | Release date: | 2023-08-23 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The S1'-S3' Pocket of the SARS-CoV-2 Main Protease Is Critical for Substrate Selectivity and Can Be Targeted with Covalent Inhibitors.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
2YSW
 
 | | Crystal Structure of the 3-dehydroquinate dehydratase from Aquifex aeolicus VF5 | | Descriptor: | 3-dehydroquinate dehydratase | | Authors: | Tanaka, T, Kumarevel, T.S, Ebihara, A, Chen, L, Fu, Z.Q, Chrzas, J, Wang, B.C, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-04 | | Release date: | 2007-10-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of the 3-dehydroquinate dehydratase from Aquifex aeolicus VF5
To be Published
|
|
7UR5
 
 | | allo-tRNAUTu1 in the A, P, and E sites of the E. coli ribosome | | Descriptor: | allo-tRNAUTu1 | | Authors: | Zhang, J, Krahn, N, Prabhakar, A, Vargas-Rodriguez, O, Krupkin, M, Fu, Z, Acosta-Reyes, F.J, Ge, X, Choi, J, Crnkovic, A, Ehrenberg, M, Viani Puglisi, E, Soll, D, Puglisi, J. | | Deposit date: | 2022-04-21 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Uncovering translation roadblocks during the development of a synthetic tRNA.
Nucleic Acids Res., 50, 2022
|
|
7URM
 
 | | allo-tRNAUTu1A in the P site of the E. coli ribosome | | Descriptor: | allo-tRNAUTU1A | | Authors: | Zhang, J, Prabhakar, A, Krahn, N, Vargas-Rodriguez, O, Krupkin, M, Fu, Z, Acosta-Reyes, F.J, Ge, X, Choi, J, Crnkovic, A, Ehrenberg, M, Viani Puglisi, E, Soll, D, Puglisi, J. | | Deposit date: | 2022-04-22 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Uncovering translation roadblocks during the development of a synthetic tRNA.
Nucleic Acids Res., 50, 2022
|
|
7URI
 
 | | allo-tRNAUTu1A in the A site of the E. coli ribosome | | Descriptor: | Allo-tRNAUTu1A | | Authors: | Zhang, J, Prabhakar, A, Krahn, N, Vargas-Rodriguez, O, Krupkin, M, Fu, Z, Acosta-Reyes, F.J, Ge, X, Choi, J, Crnkovic, A, Ehrenberg, M, Viani Puglisi, E, Soll, D, Puglisi, J. | | Deposit date: | 2022-04-22 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Uncovering translation roadblocks during the development of a synthetic tRNA.
Nucleic Acids Res., 50, 2022
|
|
