5WC7
 
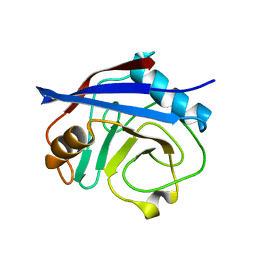 | | CypA Mutant - I97V S99T C115S | | 分子名称: | Peptidyl-prolyl cis-trans isomerase A | | 著者 | Fraser, J.S. | | 登録日 | 2017-06-29 | | 公開日 | 2018-04-18 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.43 Å) | | 主引用文献 | Rescue of conformational dynamics in enzyme catalysis by directed evolution.
Nat Commun, 9, 2018
|
|
7RGR
 
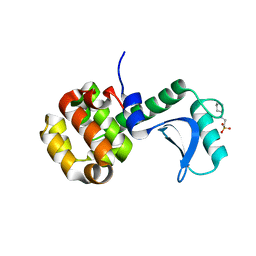 | | Lysozyme 056 from Deep neural language modeling | | 分子名称: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Artificial protein L056, CHLORIDE ION | | 著者 | Fraser, J.S, Holton, J.M, Olmos Jr, J.L, Greene, E.R. | | 登録日 | 2021-07-15 | | 公開日 | 2021-07-28 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.48 Å) | | 主引用文献 | Large language models generate functional protein sequences across diverse families.
Nat.Biotechnol., 2023
|
|
5F66
 
 | |
3K0M
 
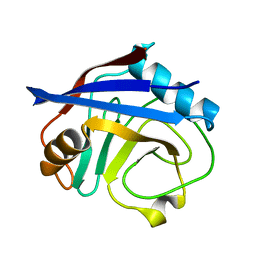 | | Cryogenic structure of CypA | | 分子名称: | Cyclophilin A | | 著者 | Fraser, J.S, Alber, T. | | 登録日 | 2009-09-24 | | 公開日 | 2009-12-08 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Hidden alternative structures of proline isomerase essential for catalysis.
Nature, 462, 2009
|
|
3K0R
 
 | |
3K0P
 
 | |
3K0N
 
 | |
3K0Q
 
 | |
3K0O
 
 | |
2NT3
 
 | | Receiver domain from Myxococcus xanthus social motility protein FrzS (Y102A Mutant) | | 分子名称: | Response regulator homolog | | 著者 | Fraser, J.S, Echols, N, Merlie, J.P, Zusman, D.R, Alber, T. | | 登録日 | 2006-11-06 | | 公開日 | 2007-03-13 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | An atypical receiver domain controls the dynamic polar localization of the Myxococcus xanthus social motility protein FrzS.
Mol.Microbiol., 65, 2007
|
|
3TGP
 
 | | Room temperature H-ras | | 分子名称: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | 著者 | Fraser, J.S, Alber, T. | | 登録日 | 2011-08-17 | | 公開日 | 2011-10-12 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.3075 Å) | | 主引用文献 | Accessing protein conformational ensembles using room-temperature X-ray crystallography.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6BTA
 
 | | CypA Mutant - S99T C115S | | 分子名称: | Peptidyl-prolyl cis-trans isomerase A | | 著者 | Fraser, J.S, Kenner, L.R, Liu, L. | | 登録日 | 2017-12-06 | | 公開日 | 2018-04-18 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Rescue of conformational dynamics in enzyme catalysis by directed evolution.
Nat Commun, 9, 2018
|
|
4OBV
 
 | | Ruminococcus gnavus tryptophan decarboxylase RUMGNA_01526 (alpha-FMT) | | 分子名称: | Pyridoxal-dependent decarboxylase domain protein, alpha-(fluoromethyl)-D-tryptophan, {5-hydroxy-4-[(1E)-4-(1H-indol-3-yl)-3-oxobut-1-en-1-yl]-6-methylpyridin-3-yl}methyl dihydrogen phosphate | | 著者 | Fraser, J.S, Van Benschoten, A.H. | | 登録日 | 2014-01-07 | | 公開日 | 2014-10-29 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.84 Å) | | 主引用文献 | Discovery and Characterization of Gut Microbiota Decarboxylases that Can Produce the Neurotransmitter Tryptamine.
Cell Host Microbe, 16, 2014
|
|
7ZJ3
 
 | | Structure of TRIM2 RING domain in complex with UBE2D1~Ub conjugate | | 分子名称: | Polyubiquitin-C, Tripartite motif-containing protein 2, Ubiquitin-conjugating enzyme E2 D1, ... | | 著者 | Esposito, D, Garza-Garcia, A, Dudley-Fraser, J, Rittinger, K. | | 登録日 | 2022-04-08 | | 公開日 | 2022-11-30 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.53 Å) | | 主引用文献 | Divergent self-association properties of paralogous proteins TRIM2 and TRIM3 regulate their E3 ligase activity.
Nat Commun, 13, 2022
|
|
2GKG
 
 | | Receiver domain from Myxococcus xanthus social motility protein FrzS | | 分子名称: | response regulator homolog | | 著者 | Echols, N, Fraser, J, Merlie, J, Zusman, D, Alber, T. | | 登録日 | 2006-04-01 | | 公開日 | 2007-03-13 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | An atypical receiver domain controls the dynamic polar localization of the Myxococcus xanthus social motility protein FrzS.
Mol.Microbiol., 65, 2007
|
|
4PTH
 
 | | Ensemble model for Escherichia coli dihydrofolate reductase at 100K | | 分子名称: | Dihydrofolate reductase, FOLIC ACID, MANGANESE (II) ION, ... | | 著者 | Keedy, D.A, van den Bedem, H, Sivak, D.A, Petsko, G.A, Ringe, D, Wilson, M.A, Fraser, J.S. | | 登録日 | 2014-03-10 | | 公開日 | 2014-05-14 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (0.85 Å) | | 主引用文献 | Crystal Cryocooling Distorts Conformational Heterogeneity in a Model Michaelis Complex of DHFR.
Structure, 22, 2014
|
|
4PST
 
 | | Multiconformer model for Escherichia coli dihydrofolate reductase at 277 K | | 分子名称: | Dihydrofolate reductase, FOLIC ACID, MANGANESE (II) ION, ... | | 著者 | Keedy, D.A, van den Bedem, H, Sivak, D.A, Petsko, G.A, Ringe, D, Wilson, M.A, Fraser, J.S. | | 登録日 | 2014-03-07 | | 公開日 | 2014-06-04 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Crystal Cryocooling Distorts Conformational Heterogeneity in a Model Michaelis Complex of DHFR.
Structure, 22, 2014
|
|
4PSS
 
 | | Multiconformer model for Escherichia coli dihydrofolate reductase at 100K | | 分子名称: | Dihydrofolate reductase, FOLIC ACID, MANGANESE (II) ION, ... | | 著者 | Keedy, D.A, van den Bedem, H, Sivak, D.A, Petsko, G.A, Ringe, D, Wilson, M.A, Fraser, J.S. | | 登録日 | 2014-03-07 | | 公開日 | 2014-06-04 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (0.849 Å) | | 主引用文献 | Crystal Cryocooling Distorts Conformational Heterogeneity in a Model Michaelis Complex of DHFR.
Structure, 22, 2014
|
|
4PTJ
 
 | | Ensemble model for Escherichia coli dihydrofolate reductase at 277K | | 分子名称: | Dihydrofolate reductase, FOLIC ACID, MANGANESE (II) ION, ... | | 著者 | Keedy, D.A, van den Bedem, H, Sivak, D.A, Petsko, G.A, Ringe, D, Wilson, M.A, Fraser, J.S. | | 登録日 | 2014-03-10 | | 公開日 | 2014-05-14 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Crystal Cryocooling Distorts Conformational Heterogeneity in a Model Michaelis Complex of DHFR.
Structure, 22, 2014
|
|
6DMK
 
 | | A multiconformer ligand model of an isoxazolyl-benzimidazole ligand bound to the bromodomain of human CREBBP | | 分子名称: | 1,2-ETHANEDIOL, 5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[2-(morpholin-4-yl)ethyl]-2-(2-phenylethyl)-1H-benzimidazole, CREB-binding protein, ... | | 著者 | Hudson, B.M, van Zundert, G, Keedy, D.A, Fonseca, R, Heliou, A, Suresh, P, Borrelli, K, Day, T, Fraser, J.S, van den Bedem, H. | | 登録日 | 2018-06-05 | | 公開日 | 2018-12-19 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | qFit-ligand Reveals Widespread Conformational Heterogeneity of Drug-Like Molecules in X-Ray Electron Density Maps.
J. Med. Chem., 61, 2018
|
|
6DMH
 
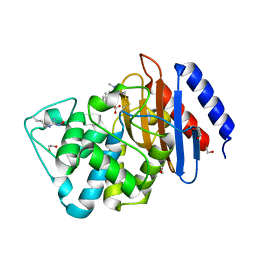 | | A multiconformer ligand model of acylenzyme intermediate of meropenem bound to an SFC-1 E166A mutant | | 分子名称: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase | | 著者 | Hudson, B.M, van Zundert, G, Keedy, D.A, Fonseca, R, Heliou, A, Suresh, P, Borrelli, K, Day, T, Fraser, J.S, van den Bedem, H. | | 登録日 | 2018-06-05 | | 公開日 | 2018-12-19 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | qFit-ligand Reveals Widespread Conformational Heterogeneity of Drug-Like Molecules in X-Ray Electron Density Maps.
J. Med. Chem., 61, 2018
|
|
6WYV
 
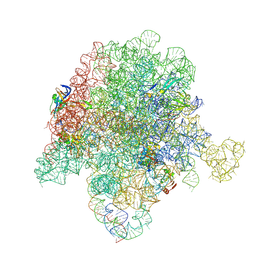 | | E. coli 50S ribosome bound to compounds 47 and VS1 | | 分子名称: | (3R,4R,5E,10E,12E,14S,16R,26aR)-16-fluoro-14-hydroxy-12-methyl-3-(propan-2-yl)-4-(prop-2-en-1-yl)-3,4,8,9,14,15,16,17,24,25,26,26a-dodecahydro-1H,7H,22H-21,18-(azeno)pyrrolo[2,1-c][1,8,4,19]dioxadiazacyclotetracosine-1,7,22-trione, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | 著者 | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | 登録日 | 2020-05-13 | | 公開日 | 2020-06-17 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (2.75 Å) | | 主引用文献 | Synthetic group A streptogramin antibiotics that overcome Vat resistance.
Nature, 586, 2020
|
|
7S1G
 
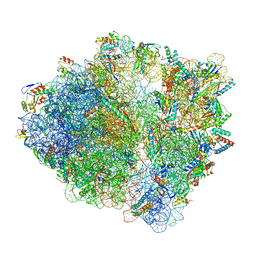 | | wild-type Escherichia coli stalled ribosome with antibiotic linezolid | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Young, I.D, Stojkovic, V, Tsai, K, Lee, D.J, Fraser, J.S, Galonic Fujimori, D. | | 登録日 | 2021-09-02 | | 公開日 | 2021-11-17 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (2.48 Å) | | 主引用文献 | Structural basis for context-specific inhibition of translation by oxazolidinone antibiotics.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7S1H
 
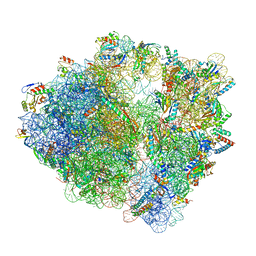 | | Wild-type Escherichia coli ribosome with antibiotic linezolid | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Young, I.D, Stojkovic, V, Tsai, K, Lee, D.J, Fraser, J.S, Galonic Fujimori, D. | | 登録日 | 2021-09-02 | | 公開日 | 2021-11-17 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (2.35 Å) | | 主引用文献 | Structural basis for context-specific inhibition of translation by oxazolidinone antibiotics.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7S1K
 
 | | Cfr-modified Escherichia coli stalled ribosome with antibiotic radezolid | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Tsai, K, Stojkovic, V, Lee, D.J, Young, I.D, Szal, T, Vazquez-Laslop, N, Mankin, A.S, Fraser, J.S, Galonic Fujimori, D. | | 登録日 | 2021-09-02 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (2.42 Å) | | 主引用文献 | Structural basis for context-specific inhibition of translation by oxazolidinone antibiotics.
Nat.Struct.Mol.Biol., 29, 2022
|
|
