8UDV
 
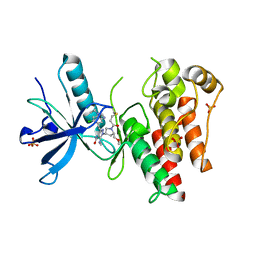 | | The X-RAY co-crystal structure of human FGFR3 V555M and Compound 17 | | Descriptor: | 1,2-ETHANEDIOL, 3-[(6-chloro-1-cyclopropyl-1H-benzimidazol-5-yl)ethynyl]-1-[(3S,5S)-5-(methoxymethyl)-1-(prop-2-enoyl)pyrrolidin-3-yl]-5-(methylamino)-1H-pyrazole-4-carboxamide, Fibroblast growth factor receptor 3, ... | | Authors: | Tyhonas, J.S, Arnold, L.D, Cox, J, Franovic, A, Gardiner, E, Grandinetti, K, Kania, R, Kanouni, T, Lardy, M, Li, C, Martin, E.S, Miller, N, Mohan, A, Murphy, E.A, Perez, M, Soroceanu, L, Timple, N, Uryu, S, Womble, S, Kaldor, S.W. | | Deposit date: | 2023-09-29 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.348 Å) | | Cite: | Discovery of KIN-3248, An Irreversible, Next Generation FGFR Inhibitor for the Treatment of Advanced Tumors Harboring FGFR2 and/or FGFR3 Gene Alterations.
J.Med.Chem., 67, 2024
|
|
8UDU
 
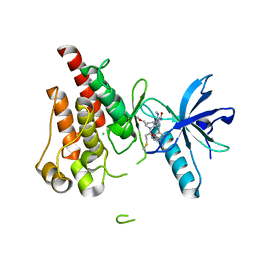 | | The X-RAY co-crystal structure of human FGFR3 and Compound 17 | | Descriptor: | 3-[(6-chloro-1-cyclopropyl-1H-benzimidazol-5-yl)ethynyl]-1-[(3S,5S)-5-(methoxymethyl)-1-(prop-2-enoyl)pyrrolidin-3-yl]-5-(methylamino)-1H-pyrazole-4-carboxamide, CHLORIDE ION, Fibroblast growth factor receptor 3 | | Authors: | Tyhonas, J.S, Arnold, L.D, Cox, J, Franovic, A, Gardiner, E, Grandinetti, K, Kania, R, Kanouni, T, Lardy, M, Li, C, Martin, E.S, Miller, N, Mohan, A, Murphy, E.A, Perez, M, Soroceanu, L, Timple, N, Uryu, S, Womble, S, Kaldor, S.W. | | Deposit date: | 2023-09-29 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.737 Å) | | Cite: | Discovery of KIN-3248, An Irreversible, Next Generation FGFR Inhibitor for the Treatment of Advanced Tumors Harboring FGFR2 and/or FGFR3 Gene Alterations.
J.Med.Chem., 67, 2024
|
|
8UDT
 
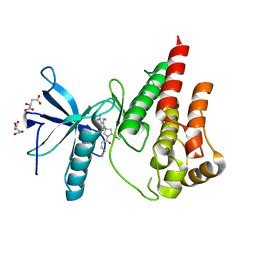 | | The X-RAY co-crystal structure of human FGFR3 and KIN-3248 | | Descriptor: | 3-[(1-cyclopropyl-4,6-difluoro-1H-benzimidazol-5-yl)ethynyl]-1-[(3R,5R)-5-(methoxymethyl)-1-propanoylpyrrolidin-3-yl]-5-(methylamino)-1H-pyrazole-4-carboxamide, D-MALATE, Fibroblast growth factor receptor 3 | | Authors: | Tyhonas, J.S, Arnold, L.D, Cox, J, Franovic, A, Gardiner, E, Grandinetti, K, Kania, R, Kanouni, T, Lardy, M, Li, C, Martin, E.S, Miller, N, Mohan, A, Murphy, E.A, Perez, M, Soroceanu, L, Timple, N, Uryu, S, Womble, S, Kaldor, S.W. | | Deposit date: | 2023-09-29 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.829 Å) | | Cite: | Discovery of KIN-3248, An Irreversible, Next Generation FGFR Inhibitor for the Treatment of Advanced Tumors Harboring FGFR2 and/or FGFR3 Gene Alterations.
J.Med.Chem., 67, 2024
|
|
3BNA
 
 | |
1MZ6
 
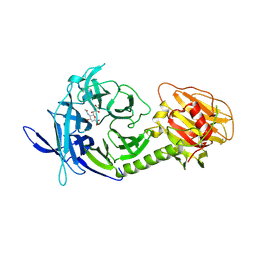 | | Trypanosoma rangeli sialidase in complex with the inhibitor DANA | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, sialidase | | Authors: | Buschiazzo, A, Tavares, G.A, Campetella, O, Spinelli, S, Cremona, M.L, Paris, G, Amaya, M.F, Frasch, A.C.C, Alzari, P.M. | | Deposit date: | 2002-10-05 | | Release date: | 2002-10-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of sialyltransferase activity in trypanosomal sialidases
Embo J., 19, 2000
|
|
1MZ5
 
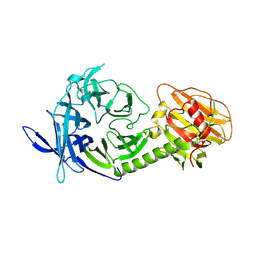 | | Trypanosoma rangeli sialidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, sialidase | | Authors: | Buschiazzo, A, Tavares, G.A, Campetella, O, Spinelli, S, Cremona, M.L, Paris, G, Amaya, M.F, Frasch, A.C.C, Alzari, P.M. | | Deposit date: | 2002-10-05 | | Release date: | 2002-10-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of sialyltransferase activity in trypanosomal sialidases
Embo J., 19, 2000
|
|
1MLX
 
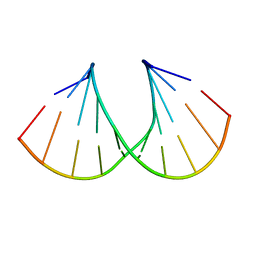 | | Crystal Structure Analysis of a 2'-O-[2-(Methylthio)-ethyl]-Modified Oligodeoxynucleotide Duplex | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*SMTP*AP*CP*GP*C)-3' | | Authors: | Prakash, T.P, Manoharan, M, Kawasaki, A.M, Fraser, A.S, Lesnik, E.A, Sioufi, N, Leeds, J.M, Teplova, M, Egli, M. | | Deposit date: | 2002-08-31 | | Release date: | 2002-12-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | 2'-O-[2-(Methylthio)ethyl]-Modified Oligonucleotide: An Analogue of 2'-O-[2-(Methoxy)-ethyl]-Modified
Oligonucleotide with Improved Protein Binding Properties and High Binding Affinity to Target RNA
Biochemistry, 41, 2002
|
|
1NMR
 
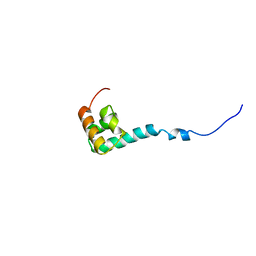 | | Solution Structure of C-terminal Domain from Trypanosoma cruzi Poly(A)-Binding Protein | | Descriptor: | poly(A)-binding protein | | Authors: | Siddiqui, N, Kozlov, G, D'Orso, I, Trempe, J.F, Frasch, A.C.C, Gehring, K. | | Deposit date: | 2003-01-10 | | Release date: | 2003-09-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the C-terminal Domain from poly(A)-binding protein in Trypanosoma cruzi: A vegetal PABC domain
Protein Sci., 12, 2003
|
|
1MS5
 
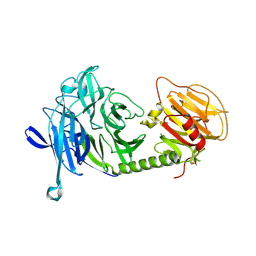 | | Triclinic form of Trypanosoma cruzi trans-sialidase, soaked with N-acetylneuraminyl-a-2,3-thio-galactoside (NA-S-Gal) | | Descriptor: | trans-sialidase | | Authors: | Buschiazzo, A, Amaya, M.F, Cremona, M.L, Frasch, A.C, Alzari, P.M. | | Deposit date: | 2002-09-19 | | Release date: | 2003-03-25 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure and mode of action of trans-sialidase, a key enzyme in Trypanosoma cruzi pathogenesis
Mol.Cell, 10, 2002
|
|
1MS0
 
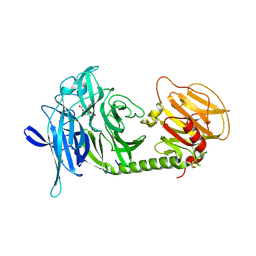 | | Monoclinic form of Trypanosoma cruzi trans-sialidase, in complex with 3-deoxy-2,3-dehydro-N-acetylneuraminic acid (DANA)and lactose | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, trans-sialidase | | Authors: | Buschiazzo, A, Amaya, M.F, Cremona, M.L, Frasch, A.C, Alzari, P.M. | | Deposit date: | 2002-09-19 | | Release date: | 2003-03-25 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure and mode of action of trans-sialidase, a key enzyme in Trypanosoma cruzi pathogenesis
Mol.Cell, 10, 2002
|
|
1MS3
 
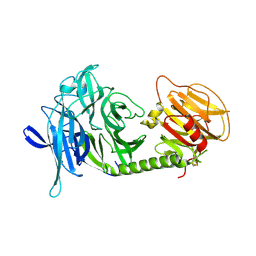 | | Monoclinic form of Trypanosoma cruzi trans-sialidase | | Descriptor: | trans-sialidase | | Authors: | Buschiazzo, A, Amaya, M.F, Cremona, M.L, Frasch, A.C, Alzari, P.M. | | Deposit date: | 2002-09-19 | | Release date: | 2003-03-25 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The crystal structure and mode of action of trans-sialidase, a key enzyme in Trypanosoma cruzi pathogenesis
Mol.Cell, 10, 2002
|
|
1MS4
 
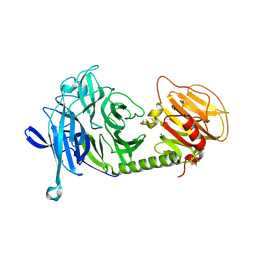 | | Triclinic form of Trypanosoma cruzi trans-sialidase | | Descriptor: | trans-sialidase | | Authors: | Buschiazzo, A, Amaya, M.F, Cremona, M.L, Frasch, A.C, Alzari, P.M. | | Deposit date: | 2002-09-19 | | Release date: | 2003-03-25 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | The crystal structure and mode of action of trans-sialidase, a key enzyme in Trypanosoma cruzi pathogenesis
Mol.Cell, 10, 2002
|
|
1MS9
 
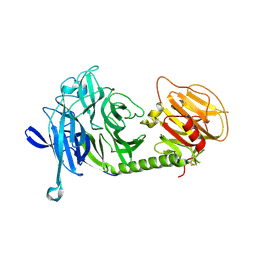 | | Triclinic form of Trypanosoma cruzi trans-sialidase, in complex with lactose | | Descriptor: | beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, trans-sialidase | | Authors: | Buschiazzo, A, Amaya, M.F, Cremona, M.L, Frasch, A.C, Alzari, P.M. | | Deposit date: | 2002-09-19 | | Release date: | 2003-03-25 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The crystal structure and mode of action of trans-sialidase, a key enzyme of Trypanosoma cruzi pathogenesis
Mol.Cell, 10, 2002
|
|
4BNA
 
 | |
1MS1
 
 | | Monoclinic form of Trypanosoma cruzi trans-sialidase, in complex with 3-deoxy-2,3-dehydro-N-acetylneuraminic acid (DANA) | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, GLYCEROL, trans-sialidase | | Authors: | Buschiazzo, A, Amaya, M.F, Cremona, M.L, Frasch, A.C, Alzari, P.M. | | Deposit date: | 2002-09-19 | | Release date: | 2003-03-25 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure and mode of action of trans-sialidase, a key enzyme in Trypanosoma cruzi pathogenesis
Mol.Cell, 10, 2002
|
|
1MR5
 
 | | Orthorhombic form of Trypanosoma cruzi trans-sialidase | | Descriptor: | trans-sialidase | | Authors: | Buschiazzo, A, Amaya, M.F, Cremona, M.L, Frasch, A.C, Alzari, P.M. | | Deposit date: | 2002-09-18 | | Release date: | 2003-03-18 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The crystal structure and mode of action of trans-sialidase, a key enzyme in Trypanosoma cruzi pathogenesis
Mol.Cell, 10, 2002
|
|
1MS8
 
 | | Triclinic form of Trypanosoma cruzi trans-sialidase, in complex with 3-deoxy-2,3-dehydro-N-acetylneuraminic acid (DANA) | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, trans-sialidase | | Authors: | Buschiazzo, A, Amaya, M.F, Cremona, M.L, Frasch, A.C, Alzari, P.M. | | Deposit date: | 2002-09-19 | | Release date: | 2003-03-25 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure and mode of action of trans-sialidase, a key enzyme in Trypanosoma cruzi pathogenesis
Mol.Cell, 10, 2002
|
|
1MNB
 
 | | BIV TAT PEPTIDE (RESIDUES 68-81), NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | BIV TAR RNA, BIV TAT PEPTIDE | | Authors: | Puglisi, J.D, Chen, L, Blanchard, S, Frankel, A.D. | | Deposit date: | 1996-07-25 | | Release date: | 1997-01-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a bovine immunodeficiency virus Tat-TAR peptide-RNA complex.
Science, 270, 1995
|
|
6QDW
 
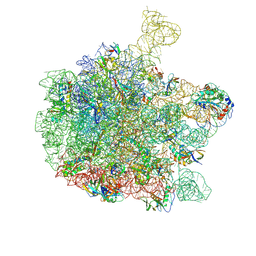 | | Cryo-EM structure of the 50S ribosomal subunit at 2.83 Angstroms with modeled GBC SecM peptide | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Schulte, L, Reitz, J, Hodirnau, V.V, Kudlinzki, D, Mao, J, Glaubitz, C, Frangakis, A, Schwalbe, H. | | Deposit date: | 2019-01-03 | | Release date: | 2020-01-15 | | Last modified: | 2020-12-02 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Cysteine oxidation and disulfide formation in the ribosomal exit tunnel.
Nat Commun, 11, 2020
|
|
4CSD
 
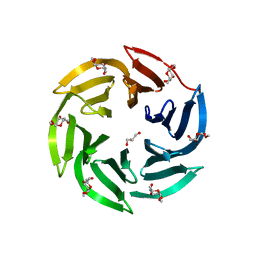 | | Structure of Monomeric Ralstonia solanacearum lectin | | Descriptor: | FUCOSE-BINDING LECTIN PROTEIN, GLYCEROL, methyl alpha-L-fucopyranoside | | Authors: | Arnaud, J, Trundle, K, Claudinon, J, Audfray, A, Varrot, A, Romer, W, Imberty, A. | | Deposit date: | 2014-03-06 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Membrane Deformation by Neolectins with Engineered Glycolipid Binding Sites.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
1ZBN
 
 | |
4LUZ
 
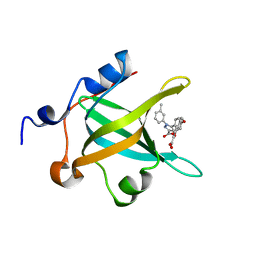 | | Fragment-Based Discovery of a Potent Inhibitor of Replication Protein A Protein-Protein Interactions | | Descriptor: | 5-(4-{[4-(5-carboxyfuran-2-yl)benzyl]oxy}phenyl)-1-(3-methylphenyl)-1H-pyrazole-3-carboxylic acid, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Frank, A.O, Kennedy, J.P, Waterson, A.G, Olejnczak, E.O, Pelz, N.F, Patrone, J.D, Vangamudi, B, Camper, D.V, Rossanese, O.W, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2013-07-25 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of a potent inhibitor of replication protein a protein-protein interactions using a fragment-linking approach.
J.Med.Chem., 56, 2013
|
|
4NB3
 
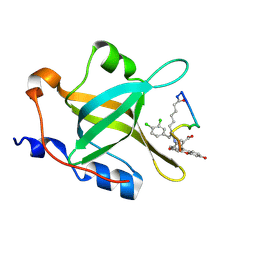 | | Crystal structure of RPA70N in complex with a 3,4 dichlorophenylalanine ATRIP derived peptide | | Descriptor: | 3,4 dichlorophenylalanine ATRIP derived peptide, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Frank, A.O, Vangamudi, B, Souza-Fagundes, E.M, Luzwik, J.W, Cortez, D, Olejniczak, O.T, Waterson, A.G, Rossanese, O.W, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2013-10-22 | | Release date: | 2014-02-26 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Discovery of a Potent Stapled Helix Peptide That Binds to the 70N Domain of Replication Protein A.
J.Med.Chem., 57, 2014
|
|
2CE6
 
 | | Structure of Helix Pomatia agglutinin with no ligands | | Descriptor: | HELIX POMATIA AGGLUTININ | | Authors: | Sanchez, J.-F, Lescar, J, Chazalet, V, Audfray, A, Gautier, C, Gagnon, J, Breton, C, Imberty, A, Mitchell, E.P. | | Deposit date: | 2006-02-03 | | Release date: | 2006-05-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Biochemical and Structural Analysis of Helix Pomatia Agglutinin. A Hexameric Lectin with a Novel Fold.
J.Biol.Chem., 281, 2006
|
|
4PMI
 
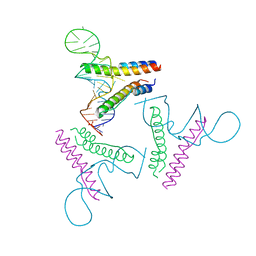 | | Crystal structure of Rev and Rev-response-element RNA complex | | Descriptor: | PHOSPHATE ION, Protein Rev, Rev-Response-Element RNA | | Authors: | Jayaraman, B, Crosby, D.C, Homer, C, Ribeiro, I, Mavor, D, Frankel, A.D. | | Deposit date: | 2014-05-21 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | RNA-directed remodeling of the HIV-1 protein Rev orchestrates assembly of the Rev-Rev response element complex.
Elife, 4, 2014
|
|
