6LO9
 
 | | Crystal structure of RORgammat with ligand C46D bound | | Descriptor: | 6-cyclohexyloxy-9-ethyl-~{N}-[(4-ethylsulfonylphenyl)methyl]carbazole-3-carboxamide, Nuclear receptor ROR-gamma | | Authors: | Feng, Y, Shijie, C. | | Deposit date: | 2020-01-04 | | Release date: | 2021-01-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.86004949 Å) | | Cite: | Crystal structure of RORgammat with ligand C46D bound
To Be Published
|
|
6LOB
 
 | | Crystal structure of RORgammat with ligand C46D bound | | Descriptor: | 9-ethyl-~{N}-[(4-ethylsulfonylphenyl)methyl]carbazole-3-carboxamide, Nuclear receptor ROR-gamma | | Authors: | Feng, Y, Shijie, C. | | Deposit date: | 2020-01-04 | | Release date: | 2021-01-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.40006661 Å) | | Cite: | Crystal structure of RORgammat with ligand C46D bound
To Be Published
|
|
6LOA
 
 | | Crystal structure of RORgammat with ligand C46D bound | | Descriptor: | 9-ethyl-~{N}-[(4-ethylsulfonylphenyl)methyl]-6-propan-2-yloxy-carbazole-3-carboxamide, Nuclear receptor ROR-gamma | | Authors: | Feng, Y, Shijie, C. | | Deposit date: | 2020-01-04 | | Release date: | 2021-01-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.50003314 Å) | | Cite: | Crystal structure of RORgammat with ligand C46D bound
To Be Published
|
|
6LOC
 
 | | Crystal structure of RORgammat with ligand C46D bound | | Descriptor: | 6-cyclobutyloxy-9-ethyl-~{N}-[(4-ethylsulfonylphenyl)methyl]carbazole-3-carboxamide, Nuclear receptor ROR-gamma | | Authors: | Feng, Y, Shijie, C. | | Deposit date: | 2020-01-04 | | Release date: | 2021-01-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.20014858 Å) | | Cite: | Crystal structure of RORgammat with ligand C46D bound
To Be Published
|
|
6LNH
 
 | | Crystal structure of IDO from Bacillus thuringiensis | | Descriptor: | FE (III) ION, L-isoleucine-4-hydroxylase, MERCURY (II) ION | | Authors: | Feng, Y, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2019-12-30 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal structure of IDO from Bacillus thuringiensis
to be published
|
|
3CVA
 
 | | Human Bcl-xL containing a Trp to Ala mutation at position 137 | | Descriptor: | Apoptosis regulator Bcl-X | | Authors: | Feng, Y, Zhang, L, Hu, T, Shen, X, Chen, K, Jiang, H, Liu, D. | | Deposit date: | 2008-04-18 | | Release date: | 2009-03-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A conserved hydrophobic core at Bcl-x(L) mediates its structural stability and binding affinity with BH3-domain peptide of pro-apoptotic protein
Arch.Biochem.Biophys., 484, 2009
|
|
6JNX
 
 | | Cryo-EM structure of a Q-engaged arrested complex | | Descriptor: | Antiterminator Q protein, DNA (63-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Feng, Y, Shi, J. | | Deposit date: | 2019-03-18 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.08 Å) | | Cite: | Structural basis of Q-dependent transcription antitermination.
Nat Commun, 10, 2019
|
|
6JNY
 
 | |
2JTM
 
 | | Solution structure of Sso6901 from Sulfolobus solfataricus P2 | | Descriptor: | Putative uncharacterized protein | | Authors: | Feng, Y, Guo, L, Huang, L, Wang, J. | | Deposit date: | 2007-08-03 | | Release date: | 2008-04-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Biochemical and structural characterization of Cren7, a novel chromatin protein conserved among Crenarchaea
Nucleic Acids Res., 36, 2008
|
|
8YLS
 
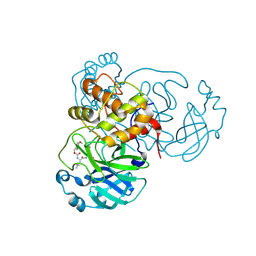 | | Structure of SARS-CoV-2 Mpro in complex with its degrader | | Descriptor: | (4-methoxyphenyl)methyl ~{N}-[(2~{S})-4-methyl-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]pentan-2-yl]carbamate, 3C-like proteinase nsp5 | | Authors: | Feng, Y, Li, W, Cheng, S.H, Li, X.B. | | Deposit date: | 2024-03-06 | | Release date: | 2024-09-04 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | The Main Protease Degraders: Potent Broad-Spectrum Agents for Anti-Coronavirus Agents
To Be Published
|
|
7XT4
 
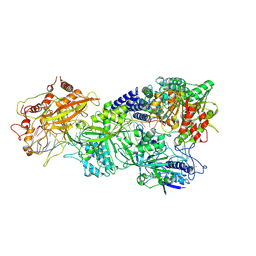 | | Structure of Craspase-NTR | | Descriptor: | CHAT domain protein, RAMP superfamily protein, RNA (34-MER), ... | | Authors: | Feng, Y, Zhang, L. | | Deposit date: | 2022-05-16 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Target RNA activates the protease activity of Craspase to confer antiviral defense.
Mol.Cell, 82, 2022
|
|
3KXT
 
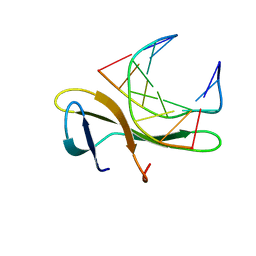 | | Crystal structure of Sulfolobus Cren7-dsDNA complex | | Descriptor: | 5'-D(*GP*CP*GP*AP*TP*CP*GP*C)-3', Chromatin protein Cren7 | | Authors: | Feng, Y, Wang, J. | | Deposit date: | 2009-12-03 | | Release date: | 2010-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Crystal structure of the crenarchaeal conserved chromatin protein Cren7 and double-stranded DNA complex
Protein Sci., 19, 2010
|
|
7XSO
 
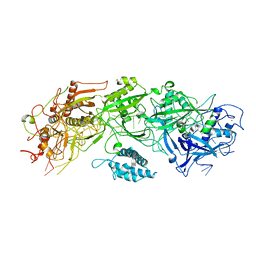 | | Structure of the type III-E CRISPR-Cas effector gRAMP | | Descriptor: | RAMP superfamily protein, RNA (35-MER), ZINC ION | | Authors: | Feng, Y, Zhang, L. | | Deposit date: | 2022-05-15 | | Release date: | 2023-03-22 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Target RNA activates the protease activity of Craspase to confer antiviral defense.
Mol.Cell, 82, 2022
|
|
7YHS
 
 | | Structure of Csy-AcrIF4-dsDNA | | Descriptor: | AcrIF4, CRISPR type I-F/YPEST-associated protein Csy2, CRISPR-associated protein Csy3, ... | | Authors: | Feng, Y, Zhang, L.X. | | Deposit date: | 2022-07-14 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Anti-CRISPR protein AcrIF4 inhibits the type I-F CRISPR-Cas surveillance complex by blocking nuclease recruitment and DNA cleavage.
J.Biol.Chem., 298, 2022
|
|
4JND
 
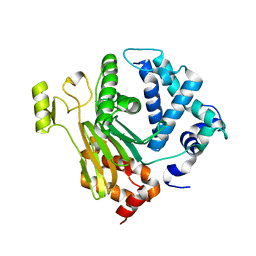 | | Structure of a C.elegans sex determining protein | | Descriptor: | Ca(2+)/calmodulin-dependent protein kinase phosphatase, MAGNESIUM ION | | Authors: | Feng, Y, Zhang, Y, Ge, J, Yang, M. | | Deposit date: | 2013-03-15 | | Release date: | 2013-06-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.652 Å) | | Cite: | Structural insight into Caenorhabditis elegans sex-determining protein FEM-2.
J.Biol.Chem., 288, 2013
|
|
1Z7R
 
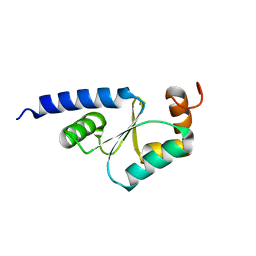 | | Solution Structure of reduced glutaredoxin C1 from Populus tremula x tremuloides | | Descriptor: | glutaredoxin | | Authors: | Feng, Y, Zhong, N, Rouhier, N, Jacquot, J.P, Xia, B. | | Deposit date: | 2005-03-26 | | Release date: | 2006-03-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Insight into Poplar Glutaredoxin C1 with a Bridging Iron-Sulfur Cluster at the Active Site
Biochemistry, 45, 2006
|
|
1Z7P
 
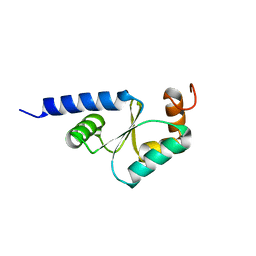 | | Solution structure of reduced glutaredoxin C1 from Populus tremula x tremuloides | | Descriptor: | glutaredoxin | | Authors: | Feng, Y, Zhong, N, Rouhier, N, Jacquot, J.P, Xia, B. | | Deposit date: | 2005-03-26 | | Release date: | 2006-03-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Insight into Poplar Glutaredoxin C1 with a Bridging Iron-Sulfur Cluster at the Active Site
Biochemistry, 45, 2006
|
|
2HR9
 
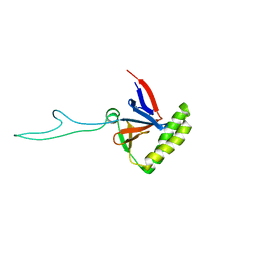 | |
6KH2
 
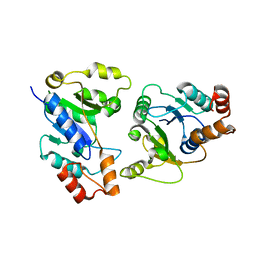 | |
2JSN
 
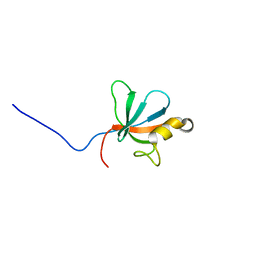 | |
6JZF
 
 | |
6JZN
 
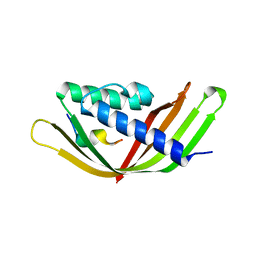 | | Structure of the intermembrane space region of PARC6-PDV1 | | Descriptor: | Peptide from Plastid division protein PDV1, Plastid division protein CDP1, chloroplastic | | Authors: | Feng, Y, Liu, Z. | | Deposit date: | 2019-05-02 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.894 Å) | | Cite: | Structure of PARC6 and PDV1 complex from Arabidopsis thaliana
To Be Published
|
|
4MEX
 
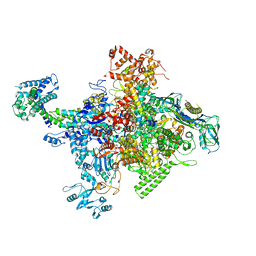 | | Crystal structure of Escherichia coli RNA polymerase in complex with salinamide A | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Feng, Y, Zhang, Y, Arnold, E, Ebright, R.H. | | Deposit date: | 2013-08-27 | | Release date: | 2014-05-21 | | Last modified: | 2014-06-25 | | Method: | X-RAY DIFFRACTION (3.902 Å) | | Cite: | Transcription inhibition by the depsipeptide antibiotic salinamide A.
Elife, 3, 2014
|
|
5HAD
 
 | |
5GTB
 
 | |
