3M5X
 
 | | Crystal structure of the mutant V182A,I199A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum | | Descriptor: | Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-03-14 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: "remote" residues that stabilize the active conformation.
Biochemistry, 49, 2010
|
|
3PNZ
 
 | | Crystal structure of the lactonase Lmo2620 from Listeria monocytogenes | | Descriptor: | GLYCEROL, PHOSPHATE ION, Phosphotriesterase family protein, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Xiang, D.F, Raushel, F.M, Almo, S.C. | | Deposit date: | 2010-11-20 | | Release date: | 2011-11-23 | | Method: | X-RAY DIFFRACTION (1.5983 Å) | | Cite: | Crystal structure of the lactonase Lmo2620 from Listeria monocytogenes
To be Published
|
|
3M3P
 
 | | Crystal structure of glutamine amido transferase from Methylobacillus Flagellatus | | Descriptor: | Glutamine amido transferase | | Authors: | Fedorov, A.A, Domagalski, M, Fedorov, E.V, Burley, S.K, Minor, W, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-03-09 | | Release date: | 2010-03-23 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of glutamine amido transferase from Methylobacillus Flagellatus
To be Published
|
|
3M43
 
 | | Crystal structure of the mutant I199A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum | | Descriptor: | GLYCEROL, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-03-10 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: "remote" residues that stabilize the active conformation.
Biochemistry, 49, 2010
|
|
3P61
 
 | | Crystal structure of the mutant R160A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, Orotidine 5'-monophosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-10-11 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.396 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: a structure-based explanation for how the 5'-phosphate group activates the enzyme.
Biochemistry, 51, 2012
|
|
3P5Z
 
 | | Crystal structure of the mutant T159S of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, GLYCEROL, N-PROPANOL, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-10-11 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: a structure-based explanation for how the 5'-phosphate group activates the enzyme.
Biochemistry, 51, 2012
|
|
3P5Y
 
 | | Crystal structure of the mutant T159A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, GLYCEROL, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-10-11 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: a structure-based explanation for how the 5'-phosphate group activates the enzyme.
Biochemistry, 51, 2012
|
|
3OZM
 
 | | Crystal structure of enolase superfamily member from Bordetella bronchiseptica complexed with Mg, m-Xylarate and L-Lyxarate | | Descriptor: | D-xylaric acid, GLYCEROL, L-arabinaric acid, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Wichelecki, D, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-09-25 | | Release date: | 2011-09-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of enolase superfamily member from Bordetella bronchiseptica complexed with Mg, m-Xylarate and L-Lyxarate
TO BE PUBLISHED
|
|
3PK7
 
 | | Crystal structure of D-mannonate dehydratase from Chromohalobacter salexigens with MG and Glycerol bound in the active site | | Descriptor: | GLYCEROL, MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme | | Authors: | Fedorov, A.A, Fedorov, E.V, Wichelecki, D, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-11-11 | | Release date: | 2011-11-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.642 Å) | | Cite: | Crystal structure of D-mannonate dehydratase from Chromohalobacter salexigens with MG and Glycerol bound in the active site
To be Published
|
|
3R25
 
 | | Crystal structure of enolase superfamily member from Vibrionales bacterium complexed with Mg and Glycerol in the active site | | Descriptor: | GLYCEROL, MAGNESIUM ION, mandelate racemase / muconate lactonizing enzyme | | Authors: | Fedorov, A.A, Fedorov, E.V, Wichelecki, D, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2011-03-13 | | Release date: | 2012-03-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.603 Å) | | Cite: | Crystal structure of enolase superfamily member from VIBRIONALES BACTERIUM complexed with Mg and Glycerol in the active site
To be Published
|
|
3RLU
 
 | | Crystal structure of the mutant K82A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with the inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, GLYCEROL, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2011-04-20 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: a structure-based explanation for how the 5'-phosphate group activates the enzyme.
Biochemistry, 51, 2012
|
|
1YDY
 
 | | Crystal structure of periplasmic glycerophosphodiester phosphodiesterase from Escherichia coli | | Descriptor: | CALCIUM ION, GLYCEROL, Glycerophosphoryl diester phosphodiesterase | | Authors: | Malashkevich, V.N, Fedorov, E, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-12-27 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of periplasmic glycerophosphodiester phosphodiesterase from Escherichia coli
To be Published
|
|
3QEZ
 
 | | Crystal structure of the mutant T159V,V182A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with the inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, GLYCEROL, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2011-01-20 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5431 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: a structure-based explanation for how the 5'-phosphate group activates the enzyme.
Biochemistry, 51, 2012
|
|
3RLV
 
 | | Crystal structure of the mutant Y206F of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with the inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, GLYCEROL, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2011-04-20 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.421 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: a structure-based explanation for how the 5'-phosphate group activates the enzyme.
Biochemistry, 51, 2012
|
|
3QMS
 
 | | Crystal structure of the mutant T159V,V182A,Y206F of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with the inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, GLYCEROL, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2011-02-05 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: a structure-based explanation for how the 5'-phosphate group activates the enzyme.
Biochemistry, 51, 2012
|
|
3QF0
 
 | | Crystal structure of the mutant T159V,Y206F of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with the inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, GLYCEROL, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2011-01-20 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: a structure-based explanation for how the 5'-phosphate group activates the enzyme.
Biochemistry, 51, 2012
|
|
1YI7
 
 | | Beta-d-xylosidase (selenomethionine) XYND from Clostridium Acetobutylicum | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-xylosidase, family 43 glycosyl hydrolase, ... | | Authors: | Teplyakov, A, Fedorov, E, Gilliland, G.L, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-01-11 | | Release date: | 2005-02-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of beta-xylosidase from clostridium acetobutylicum
To be Published
|
|
1Y7B
 
 | | BETA-D-XYLOSIDASE, A FAMILY 43 GLYCOSIDE HYDROLASE | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-xylosidase, family 43 glycosyl hydrolase, ... | | Authors: | Teplyakov, A, Fedorov, E, Gilliland, G.L, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-12-08 | | Release date: | 2005-01-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of beta-xylosidase from Clostridium acetobutylicum
To be Published
|
|
3P60
 
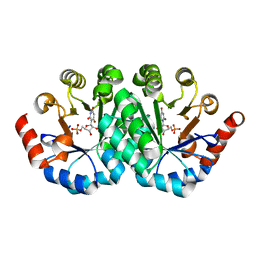 | | Crystal structure of the mutant T159V of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, Orotidine 5'-monophosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-10-11 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: a structure-based explanation for how the 5'-phosphate group activates the enzyme.
Biochemistry, 51, 2012
|
|
3OW1
 
 | | Crystal structure of D-mannonate dehydratase from Chromohalobacter salexigens complexed with MG | | Descriptor: | GLYCEROL, MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Wichelecki, D, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-09-17 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | CRYSTAL STRUCTURE OF D-MANNONATE DEHYDRATASE FROM CHROMOHALOBACTER SALEXIGENS complexed with MG
To be Published
|
|
3P93
 
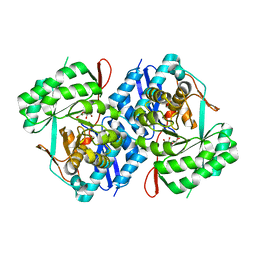 | | Crystal structure of D-mannonate dehydratase from Chromohalobacter Salexigens complexed with MG,D-Mannonate and 2-keto-3-deoxy-D-Gluconate | | Descriptor: | 2-KETO-3-DEOXYGLUCONATE, D-MANNONIC ACID, MAGNESIUM ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Wichelecki, D, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-10-15 | | Release date: | 2011-10-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CRYSTAL STRUCTURE OF D-MANNONATE DEHYDRATASE FROM CHROMOHALOBACTER SALEXIGENS complexed with MG,D-Mannonate and 2-keto-3-deoxy-D-Gluconate
To be Published
|
|
3SJ3
 
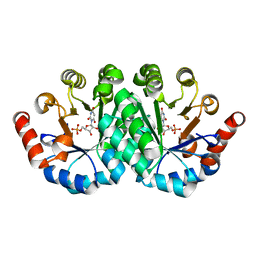 | | Crystal structure of the mutant R160A.Y206F of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with the inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2011-06-20 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: a structure-based explanation for how the 5'-phosphate group activates the enzyme.
Biochemistry, 51, 2012
|
|
3QMR
 
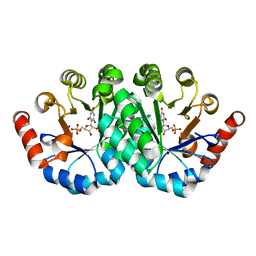 | | Crystal structure of the mutant R160A,V182A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with the inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2011-02-05 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.3213 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: a structure-based explanation for how the 5'-phosphate group activates the enzyme.
Biochemistry, 51, 2012
|
|
3QMT
 
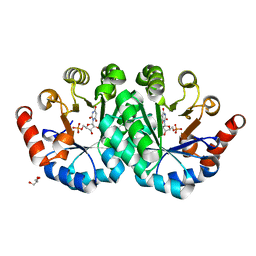 | | Crystal structure of the mutant V182A,Y206F of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with the inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, GLYCEROL, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2011-02-05 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.3202 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: a structure-based explanation for how the 5'-phosphate group activates the enzyme.
Biochemistry, 51, 2012
|
|
3O7U
 
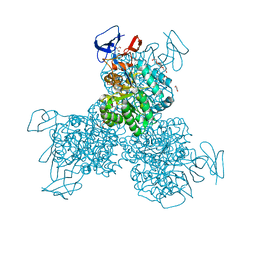 | | Crystal structure of Cytosine Deaminase from Escherichia Coli complexed with zinc and phosphono-cytosine | | Descriptor: | (2R)-2-amino-2,5-dihydro-1,5,2-diazaphosphinin-6(1H)-one 2-oxide, (2S)-1-[3-{[(2R)-2-hydroxypropyl]oxy}-2,2-bis({[(2R)-2-hydroxypropyl]oxy}methyl)propoxy]propan-2-ol, Cytosine deaminase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Hall, R.S, Raushel, F.M, Almo, S.C. | | Deposit date: | 2010-07-31 | | Release date: | 2011-06-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.708 Å) | | Cite: | Three-dimensional structure and catalytic mechanism of Cytosine deaminase.
Biochemistry, 50, 2011
|
|
