7KUO
 
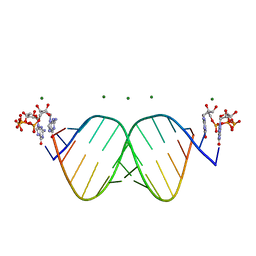 | |
7KUN
 
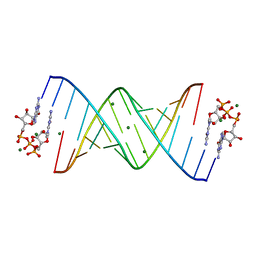 | |
7KUK
 
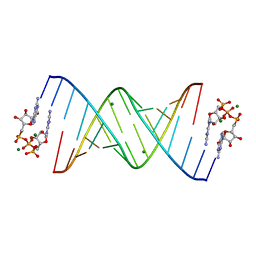 | |
7KUM
 
 | |
7KUL
 
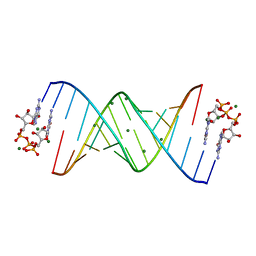 | |
7KUP
 
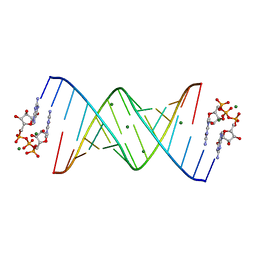 | |
7LNF
 
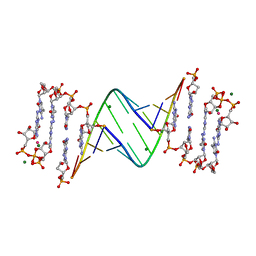 | | 3'-deoxy modification at 3' end of RNA primer complex with guanosine dinucleotide ligand G(5')ppp(5')G | | Descriptor: | 3'-deoxy-guanosine 5'-monophosphate, 4-AMINO-1-[(1S,3R,4R,7S)-7-HYDROXY-1-(HYDROXYMETHYL)-2,5-DIOXABICYCLO[2.2.1]HEPT-3-YL]-5-METHYLPYRIMIDIN-2(1H)-ONE, COBALT (II) ION, ... | | Authors: | Fang, Z, Giurgiu, C, Szostak, J.W. | | Deposit date: | 2021-02-07 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.652 Å) | | Cite: | Structure-Activity Relationships in Nonenzymatic Template-Directed RNA Synthesis.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7LNG
 
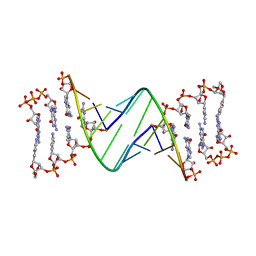 | | TNA modification at 3' end of RNA primer complex with guanosine dinucleotide ligand G(5')ppp(5')G | | Descriptor: | 2-azanyl-9-[(2~{R},3~{R},4~{S})-3-oxidanyl-4-[oxidanyl-bis(oxidanylidene)-$l^{6}-phosphanyl]oxy-oxolan-2-yl]-1~{H}-purin-6-one, 4-AMINO-1-[(1S,3R,4R,7S)-7-HYDROXY-1-(HYDROXYMETHYL)-2,5-DIOXABICYCLO[2.2.1]HEPT-3-YL]-5-METHYLPYRIMIDIN-2(1H)-ONE, DIGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Fang, Z, Giurgiu, C, Szostak, J.W. | | Deposit date: | 2021-02-07 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Activity Relationships in Nonenzymatic Template-Directed RNA Synthesis.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7LNE
 
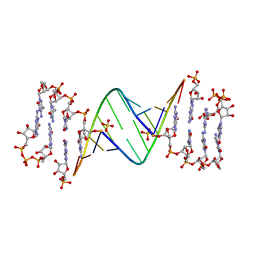 | | ANA modification at 3' end of RNA primer complex with guanosine dinucleotide ligand G(5')ppp(5')G | | Descriptor: | 4-AMINO-1-[(1S,3R,4R,7S)-7-HYDROXY-1-(HYDROXYMETHYL)-2,5-DIOXABICYCLO[2.2.1]HEPT-3-YL]-5-METHYLPYRIMIDIN-2(1H)-ONE, DIGUANOSINE-5'-TRIPHOSPHATE, GUANINE ARABINOSE-5'-PHOSPHATE, ... | | Authors: | Fang, Z, Giurgiu, C, Szostak, J.W. | | Deposit date: | 2021-02-07 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structure-Activity Relationships in Nonenzymatic Template-Directed RNA Synthesis.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6CC9
 
 | | NMR data-driven model of GTPase KRas-GMPPNP:Cmpd2 complex tethered to a nanodisc | | Descriptor: | (2R,4S)-4-[(5-bromo-1H-indole-3-carbonyl)amino]-2-[(4-chlorophenyl)methyl]piperidin-1-ium, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, ... | | Authors: | Fang, Z, Marshall, C.B, Nishikawa, T, Gossert, A.D, Jansen, J.M, Jahnke, W, Ikura, M. | | Deposit date: | 2018-02-06 | | Release date: | 2018-09-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Inhibition of K-RAS4B by a Unique Mechanism of Action: Stabilizing Membrane-Dependent Occlusion of the Effector-Binding Site.
Cell Chem Biol, 25, 2018
|
|
6CCX
 
 | | NMR data-driven model of GTPase KRas-GMPPNP:Cmpd2 complex tethered to a nanodisc | | Descriptor: | (2R,4S)-4-[(5-bromo-1H-indole-3-carbonyl)amino]-2-[(4-chlorophenyl)methyl]piperidin-1-ium, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, ... | | Authors: | Fang, Z, Marshall, C.B, Nishikawa, T, Gossert, A.D, Jansen, J.M, Jahnke, W, Ikura, M. | | Deposit date: | 2018-02-07 | | Release date: | 2018-09-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Inhibition of K-RAS4B by a Unique Mechanism of Action: Stabilizing Membrane-Dependent Occlusion of the Effector-Binding Site.
Cell Chem Biol, 25, 2018
|
|
6CCH
 
 | | NMR data-driven model of GTPase KRas-GMPPNP tethered to a nanodisc (E3 state) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Fang, Z, Marshall, C.B, Nishikawa, T, Gossert, A.D, Jansen, J.M, Jahnke, W, Ikura, M. | | Deposit date: | 2018-02-07 | | Release date: | 2018-08-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Inhibition of K-RAS4B by a Unique Mechanism of Action: Stabilizing Membrane-Dependent Occlusion of the Effector-Binding Site.
Cell Chem Biol, 25, 2018
|
|
4TTI
 
 | | Crystal structure of double mutant E. Coli purine nucleoside phosphorylase with 4 FMC molecules | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type, ... | | Authors: | Stefanic, Z, Bzowska, A. | | Deposit date: | 2014-06-21 | | Release date: | 2015-07-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystallographic snapshots of ligand binding to hexameric purine nucleoside phosphorylase and kinetic studies give insight into the mechanism of catalysis.
Sci Rep, 8, 2018
|
|
4TS3
 
 | |
4TTA
 
 | | Crystal structure of double mutant E. Coli purine nucleoside phosphorylase with 2 FMC molecules | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type, ... | | Authors: | Stefanic, Z, Bzowska, A. | | Deposit date: | 2014-06-20 | | Release date: | 2015-07-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic snapshots of ligand binding to hexameric purine nucleoside phosphorylase and kinetic studies give insight into the mechanism of catalysis.
Sci Rep, 8, 2018
|
|
4TS9
 
 | |
4TTJ
 
 | | Crystal structure of double mutant E. Coli purine nucleoside phosphorylase with 6 FMC molecules | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type, ... | | Authors: | Stefanic, Z, Bzowska, A. | | Deposit date: | 2014-06-21 | | Release date: | 2015-07-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.874 Å) | | Cite: | Crystallographic snapshots of ligand binding to hexameric purine nucleoside phosphorylase and kinetic studies give insight into the mechanism of catalysis.
Sci Rep, 8, 2018
|
|
8VAX
 
 | |
8VAW
 
 | |
1ETZ
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF AN ANTI-SWEETENER FAB, NC10.14, SHOWS THE EXTENT OF STRUCTURAL DIVERSITY IN ANTIGEN RECOGNITION BY IMMUNOGLOBULINS | | Descriptor: | FAB NC10.14 - HEAVY CHAIN, FAB NC10.14 - LIGHT CHAIN, N-(P-CYANOPHENYL)-N'-DIPHENYLMETHYL-GUANIDINE-ACETIC ACID | | Authors: | Guddat, L.W, Shan, L, Broomell, C, Ramsland, P.A, Fan, Z, Anchin, J.M, Linthicum, D.S, Edmundson, A.B. | | Deposit date: | 2000-04-13 | | Release date: | 2000-10-18 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The three-dimensional structure of a complex of a murine Fab (NC10. 14) with a potent sweetener (NC174): an illustration of structural diversity in antigen recognition by immunoglobulins.
J.Mol.Biol., 302, 2000
|
|
4KOM
 
 | | The structure of hemagglutinin from avian-origin H7N9 influenza virus in complex with avian receptor analog 3'SLNLN (NeuAcα2-3Galβ1-4GlcNAcβ1-3Galβ1-4Glc) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, Hemagglutinin HA2, ... | | Authors: | Shi, Y, Zhang, W, Wang, F, Qi, J, Song, H, Wu, Y, Gao, F, Zhang, Y, Fan, Z, Gong, W, Wang, D, Shu, Y, Wang, Y, Yan, J, Gao, G.F. | | Deposit date: | 2013-05-12 | | Release date: | 2013-11-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Structures and receptor binding of hemagglutinins from human-infecting H7N9 influenza viruses.
Science, 342, 2013
|
|
1AQK
 
 | |
3U83
 
 | | Crystal structure of nectin-1 | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, Poliovirus receptor-related protein 1 | | Authors: | Zhang, N, Yan, J, Lu, G, Guo, Z, Fan, Z, Wang, J, Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2011-10-15 | | Release date: | 2012-03-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Binding of herpes simplex virus glycoprotein D to nectin-1 exploits host cell adhesion.
Nat Commun, 2, 2011
|
|
3U82
 
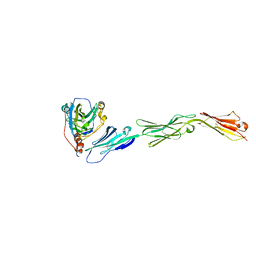 | | Binding of herpes simplex virus glycoprotein D to nectin-1 exploits host cell adhesion | | Descriptor: | Envelope glycoprotein D, Poliovirus receptor-related protein 1 | | Authors: | Zhang, N, Yan, J, Lu, G, Guo, Z, Fan, Z, Wang, J, Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2011-10-15 | | Release date: | 2012-03-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.164 Å) | | Cite: | Binding of herpes simplex virus glycoprotein D to nectin-1 exploits host cell adhesion.
Nat Commun, 2, 2011
|
|
5EAZ
 
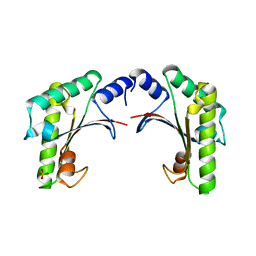 | | crystal form I of YfiB belonging to space groups P21 | | Descriptor: | SULFATE ION, YfiB | | Authors: | Xu, M, Yang, X, Yang, X.-A, Zhou, L, Liu, T.-Z, Fan, Z, Jiang, T. | | Deposit date: | 2015-10-17 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Structural insights into the regulatory mechanism of the Pseudomonas aeruginosa YfiBNR system
Protein Cell, 7, 2016
|
|
