2XIZ
 
 | | Protein kinase Pim-1 in complex with fragment-3 from crystallographic fragment screen | | Descriptor: | (E)-PYRIDIN-4-YL-ACRYLIC ACID, PROTO-ONCOGENE SERINE/THREONINE PROTEIN KINASE PIM-1 | | Authors: | Schulz, M.N, Fanghanel, J, Schafer, M, Badock, V, Briem, H, Boemer, U, Nguyen, D, Husemann, M, Hillig, R.C. | | Deposit date: | 2010-07-01 | | Release date: | 2011-02-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystallographic Fragment Screen Identifies Cinnamic Acid Derivatives as Starting Points for Potent Pim-1 Inhibitors
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2XJ1
 
 | | Protein kinase Pim-1 in complex with small molecule inibitor | | Descriptor: | (2E)-3-(3-{6-[(TRANS-4-AMINOCYCLOHEXYL)AMINO]PYRAZIN-2-YL}PHENYL)PROP-2-ENOIC ACID, PROTO-ONCOGENE SERINE/THREONINE-PROTEIN KINASE PIM-1 | | Authors: | Schulz, M.N, Fanghanel, J, Schafer, M, Badock, V, Briem, H, Boemer, U, Nguyen, D, Husemann, M, Hillig, R.C. | | Deposit date: | 2010-07-01 | | Release date: | 2011-02-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystallographic Fragment Screen Identifies Cinnamic Acid Derivatives as Starting Points for Potent Pim-1 Inhibitors
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2XIX
 
 | | Protein kinase Pim-1 in complex with fragment-1 from crystallographic fragment screen | | Descriptor: | 3,5-DIAMINO-1H-[1,2,4]TRIAZOLE, PROTO-ONCOGENE SERINE/THREONINE-PROTEIN KINASE PIM-1 | | Authors: | Schulz, M.N, Fanghanel, J, Schafer, M, Badock, V, Briem, H, Boemer, U, Nguyen, D, Husemann, M, Hillig, R.C. | | Deposit date: | 2010-07-01 | | Release date: | 2011-02-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic Fragment Screen Identifies Cinnamic Acid Derivatives as Starting Points for Potent Pim-1 Inhibitors
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2XJ2
 
 | | Protein kinase Pim-1 in complex with small molecule inhibitor | | Descriptor: | (2E)-3-{3-[6-(4-methyl-1,4-diazepan-1-yl)pyrazin-2-yl]phenyl}prop-2-enoic acid, PROTO-ONCOGENE SERINE/THREONINE-PROTEIN KINASE PIM-1 | | Authors: | Schulz, M.N, Fanghanel, J, Schafer, M, Badock, V, Briem, H, Boemer, U, Nguyen, D, Husemann, M, Hillig, R.C. | | Deposit date: | 2010-07-01 | | Release date: | 2011-02-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic Fragment Screen Identifies Cinnamic Acid Derivatives as Starting Points for Potent Pim-1 Inhibitors
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2XJ0
 
 | | Protein kinase Pim-1 in complex with fragment-4 from crystallographic fragment screen | | Descriptor: | (E)-3-(2-AMINO-PYRIDINE-5YL)-ACRYLIC ACID, PROTO-ONCOGENE SERINE/THREONINE PROTEIN KINASE PIM-1 | | Authors: | Schulz, M.N, Fanghanel, J, Schafer, M, Badock, V, Briem, H, Boemer, U, Nguyen, D, Husemann, M, Hillig, R.C. | | Deposit date: | 2010-07-01 | | Release date: | 2011-02-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystallographic Fragment Screen Identifies Cinnamic Acid Derivatives as Starting Points for Potent Pim-1 Inhibitors
Acta Crystallogr.,Sect.D, 67, 2011
|
|
7BWN
 
 | | Crystal Structure of a Designed Protein Heterocatenane | | Descriptor: | Cellular tumor antigen p53, Chimera of Green fluorescent protein and p53dim | | Authors: | Liu, Y.J, Duan, Z.L, Fang, J, Zhang, F, Xiao, J.Y, Zhang, W.B. | | Deposit date: | 2020-04-15 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | Cellular Synthesis and X-ray Crystal Structure of a Designed Protein Heterocatenane.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
2XIY
 
 | | Protein kinase Pim-1 in complex with fragment-2 from crystallographic fragment screen | | Descriptor: | 2-HYDROXYMETHYL-BENZOIMIDAZOLE, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Schulz, M.N, Fanghanel, J, Schafer, M, Badock, V, Briem, H, Boemer, U, Nguyen, D, Husemann, M, Hillig, R.C. | | Deposit date: | 2010-07-01 | | Release date: | 2011-02-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic Fragment Screen Identifies Cinnamic Acid Derivatives as Starting Points for Potent Pim-1 Inhibitors
Acta Crystallogr.,Sect.D, 67, 2011
|
|
5E5A
 
 | | Crystal structure of the chromatin-tethering domain of Human cytomegalovirus IE1 protein bound to the nucleosome core particle | | Descriptor: | C-terminal domain of Regulatory protein IE1, DNA (146-MER), Histone H2A, ... | | Authors: | Fang, Q, Chen, P, Wang, M, Fang, J, Yang, N, Li, G, Xu, R.M. | | Deposit date: | 2015-10-08 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.809 Å) | | Cite: | Human cytomegalovirus IE1 protein alters the higher-order chromatin structure by targeting the acidic patch of the nucleosome
Elife, 5, 2016
|
|
4GUS
 
 | | Crystal structure of LSD2-NPAC with H3 in space group P3221 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Histone H3.3, ... | | Authors: | Chen, F, Dong, Z, Fang, J, Yang, Y, Li, Z, Xu, Y, Yang, H, Wang, P, Fang, R, Shi, Y, Xu, Y. | | Deposit date: | 2012-08-29 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
Mol.Cell, 49, 2013
|
|
4GUR
 
 | | Crystal structure of LSD2-NPAC with H3 in space group P21 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Histone H3.3, ... | | Authors: | Chen, F, Dong, Z, Fang, J, Yang, Y, Li, Z, Xu, Y, Yang, H, Wang, P, Fang, R, Shi, Y, Xu, Y. | | Deposit date: | 2012-08-29 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
Mol.Cell, 49, 2013
|
|
4GUT
 
 | | Crystal structure of LSD2-NPAC | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Lysine-specific histone demethylase 1B, ... | | Authors: | Chen, F, Dong, Z, Fang, J, Yang, Y, Li, Z, Xu, Y, Yang, H, Wang, P, Fang, R, Shi, Y, Xu, Y. | | Deposit date: | 2012-08-29 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
Mol.Cell, 49, 2013
|
|
4GU1
 
 | | Crystal structure of LSD2 | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1B, ... | | Authors: | Chen, F, Dong, Z, Fang, J, Yang, Y, Li, Z, Xu, Y, Yang, H, Wang, P, Fang, R, Shi, Y, Xu, Y. | | Deposit date: | 2012-08-29 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.939 Å) | | Cite: | LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
Mol.Cell, 49, 2013
|
|
4BK0
 
 | | Crystal structure of the KIX domain of human RECQL5 (domain-swapped dimer) | | Descriptor: | ATP-DEPENDENT DNA HELICASE Q5, DI(HYDROXYETHYL)ETHER | | Authors: | Kassube, S.A, Jinek, M, Fang, J, Tsutakawa, S, Nogales, E. | | Deposit date: | 2013-04-21 | | Release date: | 2013-06-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Mimicry in Transcription Regulation of Human RNA Polymerase II by the DNA Helicase Recql5
Nat.Struct.Mol.Biol., 20, 2013
|
|
6MZC
 
 | | Human TFIID BC core | | Descriptor: | Transcription initiation factor TFIID subunit 10, Transcription initiation factor TFIID subunit 12, Transcription initiation factor TFIID subunit 2, ... | | Authors: | Patel, A.B, Louder, R.K, Greber, B.J, Grunberg, S, Luo, J, Fang, J, Liu, Y, Ranish, J, Hahn, S, Nogales, E. | | Deposit date: | 2018-11-05 | | Release date: | 2018-11-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure of human TFIID and mechanism of TBP loading onto promoter DNA.
Science, 362, 2018
|
|
6MZD
 
 | | Human TFIID Lobe A canonical | | Descriptor: | TATA-box-binding protein, Transcription initiation factor TFIID subunit 1, Transcription initiation factor TFIID subunit 10, ... | | Authors: | Patel, A.B, Louder, R.K, Greber, B.J, Grunberg, S, Luo, J, Fang, J, Liu, Y, Ranish, J, Hahn, S, Nogales, E. | | Deposit date: | 2018-11-05 | | Release date: | 2018-11-28 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (9.8 Å) | | Cite: | Structure of human TFIID and mechanism of TBP loading onto promoter DNA.
Science, 362, 2018
|
|
6MZM
 
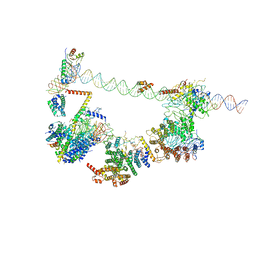 | | Human TFIID bound to promoter DNA and TFIIA | | Descriptor: | SCP DNA (80-MER), TATA-box-binding protein, Transcription initiation factor IIA subunit 1, ... | | Authors: | Patel, A.B, Louder, R.K, Greber, B.J, Grunberg, S, Luo, J, Fang, J, Liu, Y, Ranish, J, Hahn, S, Nogales, E. | | Deposit date: | 2018-11-05 | | Release date: | 2018-11-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Structure of human TFIID and mechanism of TBP loading onto promoter DNA.
Science, 362, 2018
|
|
5LIK
 
 | | Crystal structure of human AKR1B10 complexed with NADP+ and the inhibitor MK181 | | Descriptor: | 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Fanfrlik, J, Kamlar, M, Vesely, J, Hobza, P, Podjarny, A. | | Deposit date: | 2016-07-14 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | IDD388 Polyhalogenated Derivatives as Probes for an Improved Structure-Based Selectivity of AKR1B10 Inhibitors.
Acs Chem.Biol., 11, 2016
|
|
5LIY
 
 | | Crystal structure of human AKR1B10 complexed with NADP+ and the inhibitor MK204 | | Descriptor: | 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Fanfrlik, J, Kamlar, M, Vesely, J, Hobza, P, Podjarny, A. | | Deposit date: | 2016-07-15 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | IDD388 Polyhalogenated Derivatives as Probes for an Improved Structure-Based Selectivity of AKR1B10 Inhibitors.
Acs Chem.Biol., 11, 2016
|
|
5LIX
 
 | | Crystal structure of human AKR1B10 complexed with NADP+ and the inhibitor MK184 | | Descriptor: | 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Fanfrlik, J, Kamlar, M, Vesely, J, Hobza, P, Podjarny, A. | | Deposit date: | 2016-07-15 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | IDD388 Polyhalogenated Derivatives as Probes for an Improved Structure-Based Selectivity of AKR1B10 Inhibitors.
Acs Chem.Biol., 11, 2016
|
|
6MZL
 
 | | Human TFIID canonical state | | Descriptor: | TATA-box-binding protein, Transcription initiation factor TFIID subunit 1, Transcription initiation factor TFIID subunit 10, ... | | Authors: | Patel, A.B, Louder, R.K, Greber, B.J, Grunberg, S, Luo, J, Fang, J, Liu, Y, Ranish, J, Hahn, S, Nogales, E. | | Deposit date: | 2018-11-05 | | Release date: | 2018-11-28 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (23 Å) | | Cite: | Structure of human TFIID and mechanism of TBP loading onto promoter DNA.
Science, 362, 2018
|
|
5LIU
 
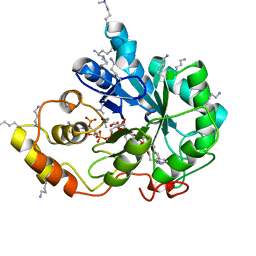 | | Crystal structure of human AKR1B10 complexed with NADP+ and the inhibitor IDD388 | | Descriptor: | (2-{[(4-BROMO-2-FLUOROBENZYL)AMINO]CARBONYL}-5-CHLOROPHENOXY)ACETIC ACID, 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member B10, ... | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Fanfrlik, J, Kamlar, M, Vesely, J, Hobza, P, Podjarny, A. | | Deposit date: | 2016-07-15 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | IDD388 Polyhalogenated Derivatives as Probes for an Improved Structure-Based Selectivity of AKR1B10 Inhibitors.
Acs Chem.Biol., 11, 2016
|
|
5LIW
 
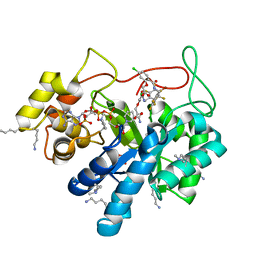 | | Crystal structure of human AKR1B10 complexed with NADP+ and the inhibitor MK319 | | Descriptor: | 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Fanfrlik, J, Kamlar, M, Vesely, J, Hobza, P, Podjarny, A. | | Deposit date: | 2016-07-15 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | IDD388 Polyhalogenated Derivatives as Probes for an Improved Structure-Based Selectivity of AKR1B10 Inhibitors.
Acs Chem.Biol., 11, 2016
|
|
4YOC
 
 | | Crystal Structure of human DNMT1 and USP7/HAUSP complex | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, Ubiquitin carboxyl-terminal hydrolase 7, ZINC ION | | Authors: | Cheng, J, Yang, H, Fang, J, Gong, R, Wang, P, Li, Z, Xu, Y. | | Deposit date: | 2015-03-11 | | Release date: | 2015-05-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.916 Å) | | Cite: | Molecular mechanism for USP7-mediated DNMT1 stabilization by acetylation.
Nat Commun, 6, 2015
|
|
5C56
 
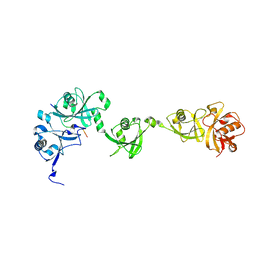 | | Crystal structure of USP7/HAUSP in complex with ICP0 | | Descriptor: | Ubiquitin E3 ligase ICP0, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Cheng, J, Li, Z, Gong, R, Fang, J, Yang, Y, Sun, C, Yang, H, Xu, Y. | | Deposit date: | 2015-06-19 | | Release date: | 2015-07-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.685 Å) | | Cite: | Molecular mechanism for the substrate recognition of USP7.
Protein Cell, 6, 2015
|
|
1L1F
 
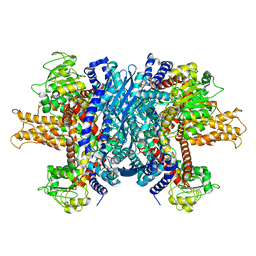 | | Structure of human glutamate dehydrogenase-apo form | | Descriptor: | Glutamate Dehydrogenase 1 | | Authors: | Smith, T.J, Schmidt, T, Fang, J, Wu, J, Siuzdak, G, Stanley, C.A. | | Deposit date: | 2002-02-15 | | Release date: | 2002-03-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of apo human glutamate dehydrogenase details subunit communication and allostery.
J.Mol.Biol., 318, 2002
|
|
