5TRS
 
 | | Structure of Mycobacterium tuberculosis proteasome in complex with N,C-capped dipeptide PKS2144 | | Descriptor: | N-tert-butoxy-N~2~-(5-methyl-1,2-oxazole-3-carbonyl)-L-asparaginyl-O-methyl-N-[(naphthalen-1-yl)methyl]-L-serinamide, Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Hsu, H.-C, Fan, H, Singh, P.K, Wang, R, Sukenick, G, Nathan, C, Lin, G, Li, H. | | Deposit date: | 2016-10-27 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.083567 Å) | | Cite: | Structural Basis for the Species-Selective Binding of N,C-Capped Dipeptides to the Mycobacterium tuberculosis Proteasome.
Biochemistry, 56, 2017
|
|
5TRR
 
 | | Structure of Mycobacterium tuberculosis proteasome in complex with N,C-capped dipeptide PKS2169 | | Descriptor: | N,N-diethyl-N~2~-(3-phenylpropanoyl)-L-asparaginyl-N-[(naphthalen-1-yl)methyl]-L-alaninamide, Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Hsu, H.-C, Fan, H, Singh, P.K, Wang, R, Sukenick, G, Nathan, C, Lin, G, Li, H. | | Deposit date: | 2016-10-27 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.103 Å) | | Cite: | Structural Basis for the Species-Selective Binding of N,C-Capped Dipeptides to the Mycobacterium tuberculosis Proteasome.
Biochemistry, 56, 2017
|
|
5TRY
 
 | | Structure of Mycobacterium tuberculosis proteasome in complex with N,C-capped dipeptide PKS2206 | | Descriptor: | (2~{S})-~{N}-[(2~{S})-3-methoxy-1-(naphthalen-1-ylmethylamino)-1-oxidanylidene-propan-2-yl]-4-oxidanylidene-2-(3-phenylpropanoylamino)-4-piperidin-1-yl-butanamide, Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Hsu, H.-C, Fan, H, Singh, P.K, Wang, R, Sukenick, G, Nathan, C, Lin, G, Li, H. | | Deposit date: | 2016-10-27 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.000008 Å) | | Cite: | Structural Basis for the Species-Selective Binding of N,C-Capped Dipeptides to the Mycobacterium tuberculosis Proteasome.
Biochemistry, 56, 2017
|
|
5TS0
 
 | | Structure of Mycobacterium tuberculosis proteasome in complex with N,C-capped dipeptide PKS2208 | | Descriptor: | (2S)-N-{(2S)-3-methoxy-1-[(naphthalen-1-ylmethyl)amino]-1-oxopropan-2-yl}-4-oxo-2-[(3-phenylpropanoyl)amino]-4-(1H-pyrrol-1-yl)butanamide (non-preferred name), Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Hsu, H.-C, Fan, H, Singh, P.K, Wang, R, Sukenick, G, Nathan, C, Lin, G, Li, H. | | Deposit date: | 2016-10-27 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.84679747 Å) | | Cite: | Structural Basis for the Species-Selective Binding of N,C-Capped Dipeptides to the Mycobacterium tuberculosis Proteasome.
Biochemistry, 56, 2017
|
|
6QWL
 
 | | Influenza B virus (B/Panama/45) polymerase Hetermotrimer in complex with 3'5' cRNA promoter | | Descriptor: | 3' cRNA, 5' cRNA, Polymerase acidic protein, ... | | Authors: | Keown, J.R, Carrique, L, Fan, H, Fodor, E, Grimes, J.M. | | Deposit date: | 2019-03-05 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structures of influenza A virus RNA polymerase offer insight into viral genome replication.
Nature, 573, 2019
|
|
2GE8
 
 | | Structure of the C-terminal dimerization domain of infectious bronchitis virus nucleocapsid protein | | Descriptor: | Nucleocapsid protein | | Authors: | Jayaram, H, Fan, H, Bowman, B.R, Ooi, A, Jayaram, J, Collisson, E.W, Lescar, J, Prasad, B.V. | | Deposit date: | 2006-03-18 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray structures of the N- and C-terminal domains of a coronavirus nucleocapsid protein: implications for nucleocapsid formation.
J.Virol., 80, 2006
|
|
2GEC
 
 | | Structure of the N-terminal domain of avian infectious bronchitis virus nucleocapsid protein (strain Gray) in a novel dimeric arrangement | | Descriptor: | Nucleocapsid protein | | Authors: | Jayaram, H, Fan, H, Bowman, B.R, Ooi, A, Jayaram, J, Collisson, E.W, Lescar, J, Prasad, B.V. | | Deposit date: | 2006-03-19 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | X-ray structures of the N- and C-terminal domains of a coronavirus nucleocapsid protein: implications for nucleocapsid formation.
J.Virol., 80, 2006
|
|
2GE7
 
 | | Structure of the C-terminal dimerization domain of infectious bronchitis virus nucleocapsid protein | | Descriptor: | Nucleocapsid protein | | Authors: | Jayaram, H, Fan, H, Bowman, B.R, Ooi, A, Jayaram, J, Collisson, E.W, Lescar, J, Prasad, B.V. | | Deposit date: | 2006-03-18 | | Release date: | 2006-06-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structures of the N- and C-terminal domains of a coronavirus nucleocapsid protein: implications for nucleocapsid formation.
J.Virol., 80, 2006
|
|
4IC5
 
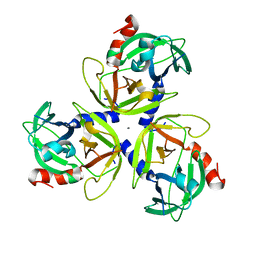 | | Crystal structure of Deg5 | | Descriptor: | CALCIUM ION, Protease Do-like 5, chloroplastic | | Authors: | Gong, W, Sun, W, Fan, H, Gao, F, Liu, L. | | Deposit date: | 2012-12-10 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.607 Å) | | Cite: | The structures of Arabidopsis Deg5 and Deg8 reveal new insights into HtrA proteases
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4IC6
 
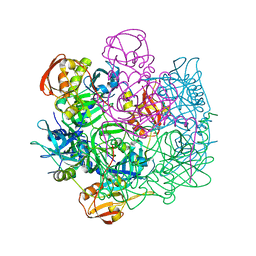 | | Crystal structure of Deg8 | | Descriptor: | Protease Do-like 8, chloroplastic | | Authors: | Gong, W, Sun, W, Fan, H, Gao, F, Liu, L. | | Deposit date: | 2012-12-10 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structures of Arabidopsis Deg5 and Deg8 reveal new insights into HtrA proteases
Acta Crystallogr.,Sect.D, 69, 2013
|
|
7EPC
 
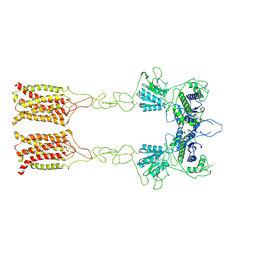 | | Cryo-EM structure of inactive mGlu7 homodimer | | Descriptor: | Isoform 3 of Metabotropic glutamate receptor 7 | | Authors: | Du, J, Wang, D, Fan, H, Tai, L, Lin, S, Han, S, Sun, F, Wu, B, Zhao, Q. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-23 | | Last modified: | 2021-07-07 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of human mGlu2 and mGlu7 homo- and heterodimers.
Nature, 594, 2021
|
|
7EPA
 
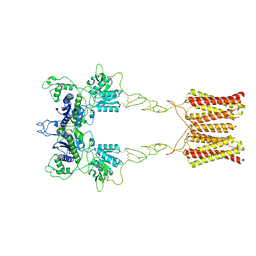 | | Cryo-EM structure of inactive mGlu2 homodimer | | Descriptor: | Metabotropic glutamate receptor 2 | | Authors: | Du, J, Wang, D, Fan, H, Tai, L, Lin, S, Han, S, Sun, F, Wu, B, Zhao, Q. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-23 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of human mGlu2 and mGlu7 homo- and heterodimers.
Nature, 594, 2021
|
|
7EPB
 
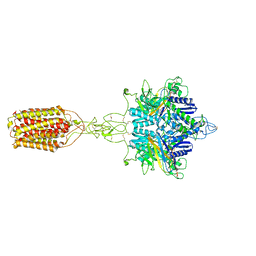 | | Cryo-EM structure of LY354740-bound mGlu2 homodimer | | Descriptor: | (1S,2S,5R,6S)-2-aminobicyclo[3.1.0]hexane-2,6-dicarboxylic acid, Anti-RON nanobody, Metabotropic glutamate receptor 2 | | Authors: | Du, J, Wang, D, Fan, H, Tai, L, Lin, S, Han, S, Sun, F, Wu, B, Zhao, Q. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-23 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of human mGlu2 and mGlu7 homo- and heterodimers.
Nature, 594, 2021
|
|
7EPD
 
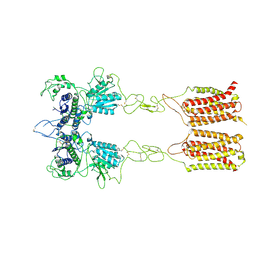 | | Cryo-EM structure of inactive mGlu2-7 heterodimer | | Descriptor: | Isoform 3 of Metabotropic glutamate receptor 7, Metabotropic glutamate receptor 2,Peptidylprolyl isomerase | | Authors: | Du, J, Wang, D, Fan, H, Tai, L, Lin, S, Han, S, Sun, F, Wu, B, Zhao, Q. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-23 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of human mGlu2 and mGlu7 homo- and heterodimers.
Nature, 594, 2021
|
|
7FHE
 
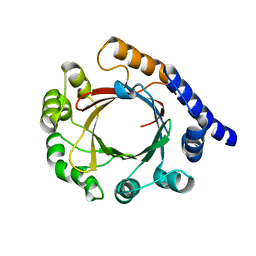 | | Structure of prenyltransferase mutant Q295F from Streptomyces sp. (strain CL190) | | Descriptor: | Prenyltransferase | | Authors: | Xue, B, Lim, K.J.H, Hartono, Y.D, Go, M.D.K, Fan, H, Yew, W.S. | | Deposit date: | 2021-07-29 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Guided Engineering of Prenyltransferase NphB for High-Yield and Regioselective Cannabinoid Production.
Acs Catalysis, 12, 2022
|
|
7FHC
 
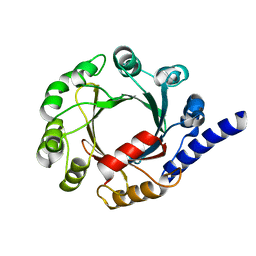 | | Structure of prenyltransferase mutant V49W from Streptomyces sp. (strain CL190) | | Descriptor: | Prenyltransferase | | Authors: | Xue, B, Lim, K.J.H, Hartono, Y.D, Go, M.D.K, Fan, H, Yew, W.S. | | Deposit date: | 2021-07-29 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structure-Guided Engineering of Prenyltransferase NphB for High-Yield and Regioselective Cannabinoid Production.
Acs Catalysis, 12, 2022
|
|
7FHB
 
 | | Structure of prenyltransferase from Streptomyces sp. (strain CL190) with bound GPP | | Descriptor: | GERANYL DIPHOSPHATE, Prenyltransferase | | Authors: | Xue, B, Lim, K.J.H, Hartono, Y.D, Go, M.D.K, Fan, H, Yew, W.S. | | Deposit date: | 2021-07-29 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Guided Engineering of Prenyltransferase NphB for High-Yield and Regioselective Cannabinoid Production.
Acs Catalysis, 12, 2022
|
|
7FHD
 
 | | Structure of prenyltransferase mutant Y288P from Streptomyces sp. (strain CL190) | | Descriptor: | Prenyltransferase | | Authors: | Xue, B, Lim, K.J.H, Hartono, Y.D, Go, M.D.K, Fan, H, Yew, W.S. | | Deposit date: | 2021-07-29 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure-Guided Engineering of Prenyltransferase NphB for High-Yield and Regioselective Cannabinoid Production.
Acs Catalysis, 12, 2022
|
|
7FHF
 
 | | Structure of prenyltransferase mutant V49W/Y288F/Q295F from Streptomyces sp. (strain CL190) | | Descriptor: | Prenyltransferase | | Authors: | Xue, B, Lim, K.J.H, Hartono, Y.D, Go, M.D.K, Fan, H, Yew, W.S. | | Deposit date: | 2021-07-29 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Guided Engineering of Prenyltransferase NphB for High-Yield and Regioselective Cannabinoid Production.
Acs Catalysis, 12, 2022
|
|
8WD5
 
 | |
