6PEY
 
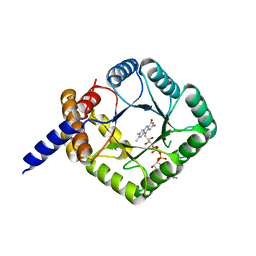 | | MTHFR with mutation Asp120Ala | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Methylenetetrahydrofolate reductase | | Authors: | Gallagher, E.L, Gurney, L.A, Frasco, F.G, Trimmer, E, Bolen, R.L, Collins, K, Garland, E, Halloran, J, Handley-Pendelton, J, Hernandez, V, Leffler, S, Perez, A, Soares, A, Stojanoff, V, Williams, D. | | Deposit date: | 2019-06-21 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Examination of Asp120Ala a Chemically Important Novel Mutation in the Enzyme Mthylenetetrahydrofolate Reductase
To be published
|
|
6PSA
 
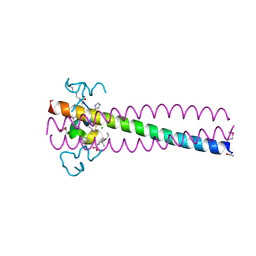 | | PIE12 D-PEPTIDE AGAINST HIV ENTRY (IN COMPLEX WITH IQN17 Q577R RESISTANCE MUTANT) | | Descriptor: | CHLORIDE ION, IQN17, PIE12 D-peptide | | Authors: | Hill, C.P, Whitby, F.G, Kay, M, Weinstock, M. | | Deposit date: | 2019-07-12 | | Release date: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Characterization of resistance to a potent D-peptide HIV entry inhibitor.
Retrovirology, 16, 2019
|
|
3UPH
 
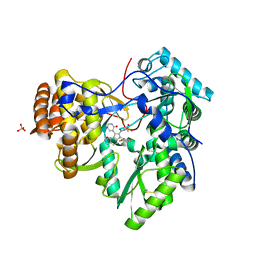 | | Synthesis of novel 4,5-dihydrofurano indoles and their evaluation as HCV NS5B polymerase inhibitors | | Descriptor: | 6-(2,5-difluorobenzyl)-N-(methylsulfonyl)-8-(2-oxo-1,2-dihydropyridin-3-yl)-3,6-dihydro-2H-furo[2,3-e]indole-7-carboxamide, PHOSPHATE ION, RNA-directed RNA polymerase | | Authors: | Velazquez, F, Venkataraman, S, Lesburg, C.A, Duca, J.S, Rosenblum, S.B, Kozlowski, J.A, Njoroge, F.G. | | Deposit date: | 2011-11-18 | | Release date: | 2012-01-25 | | Last modified: | 2012-02-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis of New 4,5-Dihydrofuranoindoles and Their Evaluation as HCV NS5B Polymerase Inhibitors.
Org.Lett., 14, 2012
|
|
3UPI
 
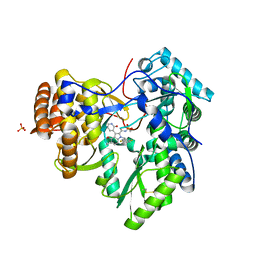 | | Synthesis of novel 4,5-dihydrofurano indoles and their evaluation as HCV NS5B polymerase inhibitors | | Descriptor: | (3S)-6-(2,5-difluorobenzyl)-3-methyl-N-(methylsulfonyl)-8-(2-oxo-1,2-dihydropyridin-3-yl)-3,6-dihydro-2H-furo[2,3-e]indole-7-carboxamide, PHOSPHATE ION, RNA-directed RNA polymerase | | Authors: | Velazquez, F, Venkataraman, S, Lesburg, C.A, Duca, J.S, Rosenblum, S.B, Kozlowski, J.A, Njoroge, F.G. | | Deposit date: | 2011-11-18 | | Release date: | 2012-01-25 | | Last modified: | 2012-02-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis of New 4,5-Dihydrofuranoindoles and Their Evaluation as HCV NS5B Polymerase Inhibitors.
Org.Lett., 14, 2012
|
|
2BLI
 
 | | L29W Mb deoxy | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nienhaus, K, Ostermann, A, Nienhaus, G.U, Parak, F.G, Schmidt, M. | | Deposit date: | 2005-03-04 | | Release date: | 2005-04-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Ligand Migration and Protein Fluctuations in Myoglobin Mutant L29W
Biochemistry, 44, 2005
|
|
2BLH
 
 | | Ligand Migration and Protein Fluctuations in Myoglobin Mutant L29W | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nienhaus, K, Ostermann, A, Nienhaus, G.U, Parak, F.G, Schmidt, M. | | Deposit date: | 2005-03-04 | | Release date: | 2005-04-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Ligand Migration and Protein Fluctuations in Myoglobin Mutant L29W
Biochemistry, 44, 2005
|
|
2BLJ
 
 | | Structure of L29W MbCO | | Descriptor: | CARBON MONOXIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nienhaus, K, Ostermann, A, Nienhaus, G.U, Parak, F.G, Schmidt, M. | | Deposit date: | 2005-03-04 | | Release date: | 2005-04-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ligand Migration and Protein Fluctuations in Myoglobin Mutant L29W
Biochemistry, 44, 2005
|
|
2D3R
 
 | | Cratylia folibunda seed lectin at acidic pH | | Descriptor: | CALCIUM ION, Lectin alpha chain, MANGANESE (II) ION | | Authors: | Del Sol, F.G, Cavada, B.S, Calvete, J.J. | | Deposit date: | 2005-09-30 | | Release date: | 2005-11-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of Cratylia floribunda seed lectin at acidic and basic pHs. Insights into the structural basis of the pH-dependent dimer-tetramer transition.
J.Struct.Biol., 158, 2007
|
|
2D3P
 
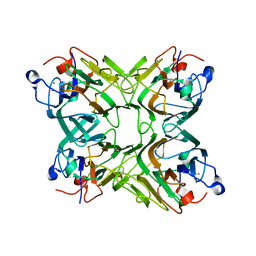 | |
2FM2
 
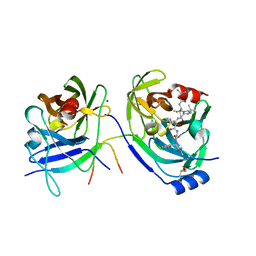 | | HCV NS3-4A protease domain complexed with a ketoamide inhibitor, SCH446211 | | Descriptor: | BETA-MERCAPTOETHANOL, NS3 protease/helicase, NS4a protein, ... | | Authors: | Yi, M, Tong, X, Skelton, A, Chase, R, Chen, T, Prongay, A, Bogen, S.L, Saksena, A.K, Njoroge, F.G, Veselenak, R.L, Pyles, R.B, Bourne, N, Malcolm, B.A, Lemon, S.M. | | Deposit date: | 2006-01-06 | | Release date: | 2006-02-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mutations conferring resistance to SCH6, a novel hepatitis C virus NS3/4A protease inhibitor. Reduced RNA replication fitness and partial rescue by second-site mutations
J.Biol.Chem., 281, 2006
|
|
1FNT
 
 | | CRYSTAL STRUCTURE OF THE 20S PROTEASOME FROM YEAST IN COMPLEX WITH THE PROTEASOME ACTIVATOR PA26 FROM TRYPANOSOME BRUCEI AT 3.2 ANGSTROMS RESOLUTION | | Descriptor: | MAGNESIUM ION, PROTEASOME ACTIVATOR PROTEIN PA26, PROTEASOME COMPONENT C1, ... | | Authors: | Whitby, F.G, Masters, E, Kramer, L, Knowlton, J.R, Yao, Y, Wang, C.C, Hill, C.P. | | Deposit date: | 2000-08-23 | | Release date: | 2001-04-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for the activation of 20S proteasomes by 11S regulators.
Nature, 408, 2000
|
|
1DO4
 
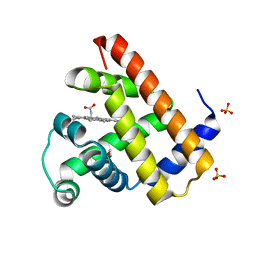 | | CARBONMONOXY-MYOGLOBIN (MUTANT L29W) AFTER PHOTOLYSIS AT T<180K | | Descriptor: | CARBON MONOXIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Ostermann, A, Waschipky, R, Parak, F.G, Nienhaus, G.U. | | Deposit date: | 1999-12-18 | | Release date: | 2000-01-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Ligand binding and conformational motions in myoglobin.
Nature, 404, 2000
|
|
1DO7
 
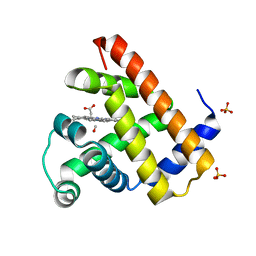 | | CARBONMONOXY-MYOGLOBIN (MUTANT L29W) REBINDING STRUCTURE AFTER PHOTOLYSIS AT T< 180K | | Descriptor: | CARBON MONOXIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Ostermann, A, Waschipky, R, Parak, F.G, Nienhaus, G.U. | | Deposit date: | 1999-12-19 | | Release date: | 2000-01-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Ligand binding and conformational motions in myoglobin.
Nature, 404, 2000
|
|
1DO1
 
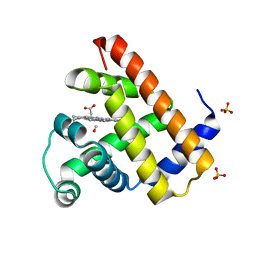 | | CARBONMONOXY-MYOGLOBIN MUTANT L29W AT 105K | | Descriptor: | CARBON MONOXIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Ostermann, A, Waschipky, R, Parak, F.G, Nienhaus, G.U. | | Deposit date: | 1999-12-18 | | Release date: | 2000-01-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Ligand binding and conformational motions in myoglobin.
Nature, 404, 2000
|
|
1H6R
 
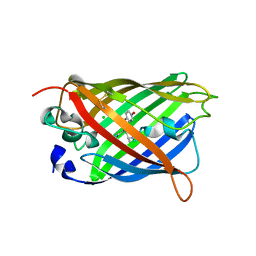 | | The oxidized state of a redox sensitive variant of green fluorescent protein | | Descriptor: | CHLORIDE ION, GREEN FLUORESCENT PROTEIN | | Authors: | Ostergaard, H, Henriksen, A, Hansen, F.G, Winther, J.R. | | Deposit date: | 2001-06-22 | | Release date: | 2001-11-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Shedding Light on Disulfide Bond Formation: Engineering a Redox Switch in Green Fluorescent Protein
Embo J., 20, 2001
|
|
1DO3
 
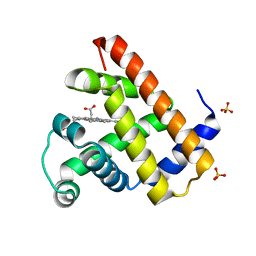 | | CARBONMONOXY-MYOGLOBIN (MUTANT L29W) AFTER PHOTOLYSIS AT T>180K | | Descriptor: | CARBON MONOXIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Ostermann, A, Waschipky, R, Parak, F.G, Nienhaus, G.U. | | Deposit date: | 1999-12-18 | | Release date: | 2000-01-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Ligand binding and conformational motions in myoglobin.
Nature, 404, 2000
|
|
1N0L
 
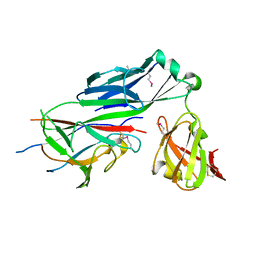 | | Crystal structure of the PapD chaperone (C-terminally 6x histidine-tagged) bound to the PapE pilus subunit (N-terminal-deleted) from uropathogenic E. coli | | Descriptor: | Chaperone protein PapD, mature Fimbrial protein PapE | | Authors: | Sauer, F.G, Pinkner, J.S, Waksman, G, Hultgren, S.J. | | Deposit date: | 2002-10-14 | | Release date: | 2002-12-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Chaperone priming of pilus subunits facilitates a topological transition that drives fiber formation
Cell(Cambridge,Mass.), 111, 2002
|
|
1NDD
 
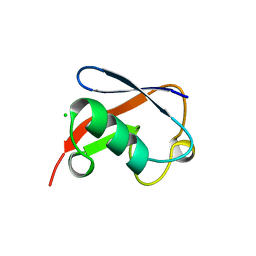 | | STRUCTURE OF NEDD8 | | Descriptor: | CHLORIDE ION, PROTEIN (UBIQUITIN-LIKE PROTEIN NEDD8), SULFATE ION | | Authors: | Whitby, F.G, Xia, G, Pickart, C.M, Hill, C.P. | | Deposit date: | 1998-08-21 | | Release date: | 1999-02-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the human ubiquitin-like protein NEDD8 and interactions with ubiquitin pathway enzymes.
J.Biol.Chem., 273, 1998
|
|
1P49
 
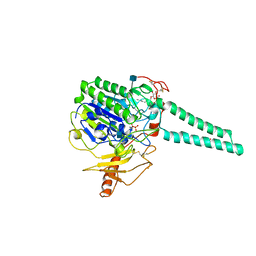 | | Structure of Human Placental Estrone/DHEA Sulfatase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, PHOSPHATE ION, ... | | Authors: | Hernandez-Guzman, F.G, Higashiyama, T, Pangborn, W, Osawa, Y, Ghosh, D. | | Deposit date: | 2003-04-21 | | Release date: | 2003-08-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Human Estrone Sulfatase Suggests Functional Roles of Membrane Association
J.Biol.Chem., 278, 2003
|
|
1PDK
 
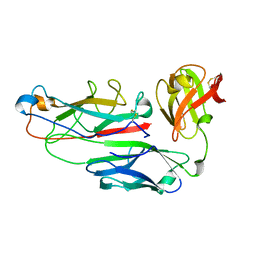 | | PAPD-PAPK CHAPERONE-PILUS SUBUNIT COMPLEX FROM E.COLI P PILUS | | Descriptor: | PROTEIN (CHAPERONE PROTEIN PAPD), PROTEIN (PROTEIN PAPK) | | Authors: | Fuetterer, K, Sauer, F.G, Hultgren, S.J, Waksman, G. | | Deposit date: | 1999-03-29 | | Release date: | 1999-08-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of chaperone function and pilus biogenesis.
Science, 285, 1999
|
|
2XSZ
 
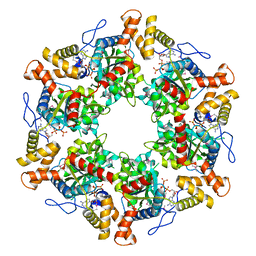 | | The dodecameric human RuvBL1:RuvBL2 complex with truncated domains II | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, RUVB-LIKE 1, RUVB-LIKE 2 | | Authors: | Gorynia, S, Bandeiras, T.M, Matias, P.M, Pinho, F.G, McVey, C.E, Vonrhein, C, Svergun, D.I, Round, A, Donner, P, Carrondo, M.A. | | Deposit date: | 2010-10-01 | | Release date: | 2011-10-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and Functional Insights Into a Dodecameric Molecular Machine - the Ruvbl1/Ruvbl2 Complex.
J.Struct.Biol., 176, 2011
|
|
1N12
 
 | | Crystal structure of the PapE (N-terminal-deleted) pilus subunit bound to a peptide corresponding to the N-terminal extension of the PapK pilus subunit (residues 1-11) from uropathogenic E. coli | | Descriptor: | Peptide corresponding to the N-terminal extension of protein PapK, mature Fimbrial protein PapE | | Authors: | Sauer, F.G, Pinkner, J.S, Waksman, G, Hultgren, S.J. | | Deposit date: | 2002-10-16 | | Release date: | 2002-12-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Chaperone priming of pilus subunits facilitates a topological transition that drives fiber formation
Cell(Cambridge,Mass.), 111, 2002
|
|
3B6H
 
 | | Crystal structure of human prostacyclin synthase in complex with inhibitor minoxidil | | Descriptor: | 6-PIPERIDIN-1-YLPYRIMIDINE-2,4-DIAMINE 3-OXIDE, PROTOPORPHYRIN IX CONTAINING FE, Prostacyclin synthase, ... | | Authors: | Li, Y.-C, Chiang, C.-W, Yeh, H.-C, Hsu, P.-Y, Whitby, F.G, Wang, L.-H, Chan, N.-L. | | Deposit date: | 2007-10-29 | | Release date: | 2007-11-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structures of Prostacyclin Synthase and Its Complexes with Substrate Analog and Inhibitor Reveal a Ligand-specific Heme Conformation Change
J.Biol.Chem., 283, 2008
|
|
3B99
 
 | | Crystal structure of zebrafish prostacyclin synthase (cytochrome P450 8A1) in complex with substrate analog U51605 | | Descriptor: | (5Z)-7-{(1R,4S,5R,6R)-6-[(1E)-oct-1-en-1-yl]-2,3-diazabicyclo[2.2.1]hept-2-en-5-yl}hept-5-enoic acid, PROTOPORPHYRIN IX CONTAINING FE, Prostaglandin I2 synthase | | Authors: | Li, Y.-C, Chiang, C.-W, Yeh, H.-C, Hsu, P.-Y, Whitby, F.G, Wang, L.-H, Chan, N.-L. | | Deposit date: | 2007-11-03 | | Release date: | 2007-11-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of Prostacyclin Synthase and Its Complexes with Substrate Analog and Inhibitor Reveal a Ligand-specific Heme Conformation Change
J.Biol.Chem., 283, 2008
|
|
3B98
 
 | | Crystal structure of zebrafish prostacyclin synthase (cytochrome P450 8A1) | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Prostaglandin I2 synthase | | Authors: | Li, Y.-C, Chiang, C.-W, Yeh, H.-C, Hsu, P.-Y, Whitby, F.G, Wang, L.-H, Chan, N.-L. | | Deposit date: | 2007-11-03 | | Release date: | 2007-11-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structures of Prostacyclin Synthase and Its Complexes with Substrate Analog and Inhibitor Reveal a Ligand-specific Heme Conformation Change
J.Biol.Chem., 283, 2008
|
|
