6WBU
 
 | |
6VPD
 
 | | Crystal structure of Trgpx in apo form | | Descriptor: | Glutathione peroxidase | | Authors: | Adriani, P.P, De Oliveira, G.S, Paiva, F.C.R, Dias, M.V.B, Chambergo, F.S. | | Deposit date: | 2020-02-03 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Structural and functional characterization of the glutathione peroxidase-like thioredoxin peroxidase from the fungus Trichoderma reesei.
Int.J.Biol.Macromol., 167, 2020
|
|
4LA7
 
 | | X-ray crystal structure of the PYL2-quinabactin-Hab1 ternary complex | | Descriptor: | ACETATE ION, Abscisic acid receptor PYL2, GLYCEROL, ... | | Authors: | Peterson, F.C, Volkman, B.F, Cutler, S.R. | | Deposit date: | 2013-06-19 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Activation of dimeric ABA receptors elicits guard cell closure, ABA-regulated gene expression, and drought tolerance.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1HQB
 
 | | TERTIARY STRUCTURE OF APO-D-ALANYL CARRIER PROTEIN | | Descriptor: | APO-D-ALANYL CARRIER PROTEIN | | Authors: | Volkman, B.F, Zhang, Q, Debabov, D.V, Rivera, E, Kresheck, G, Neuhaus, F.C. | | Deposit date: | 2000-12-14 | | Release date: | 2001-08-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Biosynthesis of D-alanyl-lipoteichoic acid: the tertiary structure of apo-D-alanyl carrier protein.
Biochemistry, 40, 2001
|
|
6FGM
 
 | | The NMR solution structure of the peptide AC12 from Hypsiboas raniceps | | Descriptor: | ALA-CYS-PHE-LEU-THR-ARG-LEU-GLY-THR-TYR-VAL-CYS | | Authors: | Popov, C.S.F.C, Simas, B.S, Goodfellow, B.J, Bocca, A.L, Andrade, P.B, Pereira, D, Valentao, P, Pereira, P.J.B, Rodrigues, J.E, Veloso Jr, P.H.H, Rezende, T.M.B. | | Deposit date: | 2018-01-11 | | Release date: | 2019-01-09 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Host-defense peptides AC12, DK16 and RC11 with immunomodulatory activity isolated from Hypsiboas raniceps skin secretion.
Peptides, 113, 2019
|
|
3LHR
 
 | | Crystal structure of the SCAN domain from Human ZNF24 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ETHYL MERCURY ION, ... | | Authors: | Volkman, B.F, Peterson, F.C, Bingman, C.A, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2010-01-22 | | Release date: | 2010-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the SCAN domain from Human ZNF24
To be Published
|
|
7XIV
 
 | | Structural insight into the interactions between Lloviu virus VP30 and nucleoprotein | | Descriptor: | Nucleocapsid protein,Minor nucleoprotein VP30 | | Authors: | Dong, S.S, Qin, X.C, Sun, W.Y, Luan, F.C, Wang, J.J, Ma, L, Li, X.X, Yang, G.X, Hao, C.Y. | | Deposit date: | 2022-04-14 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Structural insights into the interactions between lloviu virus VP30 and nucleoprotein.
Biochem.Biophys.Res.Commun., 616, 2022
|
|
7PA5
 
 | | Complex between the beta-lactamase CMY-2 with an inhibitory nanobody | | Descriptor: | Beta-lactamase, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Frederic Cawez, F.C, Frederic Kerff, F.K, Moreno Galleni, M.G, Raphael Herman, R.H. | | Deposit date: | 2021-07-29 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.184 Å) | | Cite: | Development of Nanobodies as Theranostic Agents against CMY-2-Like Class C beta-Lactamases.
Antimicrob.Agents Chemother., 67, 2023
|
|
2BE6
 
 | | 2.0 A crystal structure of the CaV1.2 IQ domain-Ca/CaM complex | | Descriptor: | CALCIUM ION, Calmodulin 2, NICKEL (II) ION, ... | | Authors: | Van Petegem, F, Chatelain, F.C, Minor Jr, D.L. | | Deposit date: | 2005-10-23 | | Release date: | 2005-11-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into voltage-gated calcium channel regulation from the structure of the Ca(V)1.2 IQ domain-Ca(2+)/calmodulin complex
Nat.Struct.Mol.Biol., 12, 2005
|
|
6S2P
 
 | |
5I7Z
 
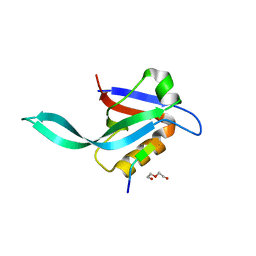 | | Crystal structure of a Par-6 PDZ-Crumbs 3 C-terminal peptide complex | | Descriptor: | Crb-3, DI(HYDROXYETHYL)ETHER, LD29223p | | Authors: | Whitney, D.S, Peterson, F.C, Prehoda, K.E, Volkman, B.F. | | Deposit date: | 2016-02-18 | | Release date: | 2016-03-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Binding of Crumbs to the Par-6 CRIB-PDZ Module Is Regulated by Cdc42.
Biochemistry, 55, 2016
|
|
7JH1
 
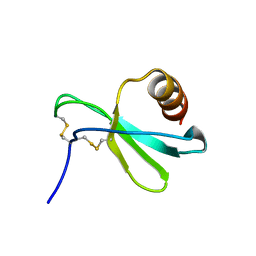 | |
8B8V
 
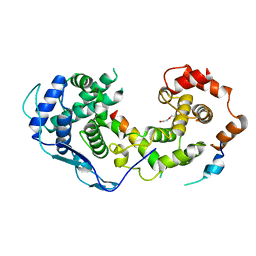 | |
9COJ
 
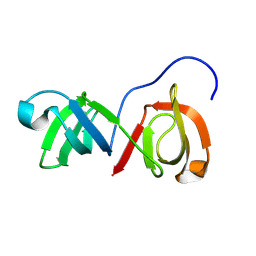 | | SH3-like tandem domain of human KIN protein | | Descriptor: | DNA/RNA-binding protein KIN17 | | Authors: | de Lourenco, I.O, Fossey, M.A, Souza, F.P, Seixas, F.A.V, Fernandez, M.A, Almeida, F.C.L, Caruso, I.P. | | Deposit date: | 2024-07-16 | | Release date: | 2024-07-31 | | Method: | SOLUTION NMR | | Cite: | Solution structure of SH3-like tandem domain of human KIN protein and its interaction with RNA homo-oligonucleotides
To Be Published
|
|
5SZW
 
 | | NMR solution structure of the RRM1 domain of the post-transcriptional regulator HuR | | Descriptor: | ELAV-like protein 1 | | Authors: | Lixa, C, Mujo, A, Jendiroba, K.A, Almeida, F.C.L, Lima, L.M.T.R, Pinheiro, A.S. | | Deposit date: | 2016-08-15 | | Release date: | 2017-09-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Oligomeric transition and dynamics of RNA binding by the HuR RRM1 domain in solution.
J. Biomol. NMR, 72, 2018
|
|
4WWC
 
 | | Crystal structure of full length YvoA in complex with palindromic operator DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(P*CP*AP*GP*TP*GP*GP*TP*CP*TP*AP*GP*AP*CP*CP*AP*CP*TP*GP*G)-3'), HTH-type transcriptional repressor YvoA | | Authors: | Grau, F.C, Fillenberg, S.B, Muller, Y.A. | | Deposit date: | 2014-11-10 | | Release date: | 2015-01-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.903 Å) | | Cite: | Structural insight into operator dre-sites recognition and effector binding in the GntR/HutC transcription regulator NagR.
Nucleic Acids Res., 43, 2015
|
|
8EOD
 
 | |
3CB7
 
 | | The crystallographic structure of the digestive lysozyme 2 from Musca domestica at 1.9 Ang. | | Descriptor: | ACETIC ACID, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Cancado, F.C, Valerio, A.A, Marana, S.R, Barbosa, J.A.R.G. | | Deposit date: | 2008-02-21 | | Release date: | 2009-02-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystallographic structure of the digestive lysozyme 2 from Musca domestica at 1.9 Ang.
To be Published
|
|
8EY0
 
 | | Structure of an orthogonal PYR1*:HAB1* chemical-induced dimerization module in complex with mandipropamid | | Descriptor: | (2S)-2-(4-chlorophenyl)-N-{2-[3-methoxy-4-(prop-2-yn-1-yloxy)phenyl]ethyl}-2-(prop-2-yn-1-yloxy)ethanamide, Abscisic acid receptor PYR1, GLYCEROL, ... | | Authors: | Park, S.-Y, Volkman, B.F, Cutler, S.R, Peterson, F.C. | | Deposit date: | 2022-10-26 | | Release date: | 2023-11-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An orthogonalized PYR1-based CID module with reprogrammable ligand-binding specificity.
Nat.Chem.Biol., 20, 2024
|
|
4WVO
 
 | | An engineered PYR1 mandipropamid receptor in complex with mandipropamid and HAB1 | | Descriptor: | (2S)-2-(4-chlorophenyl)-N-{2-[3-methoxy-4-(prop-2-yn-1-yloxy)phenyl]ethyl}-2-(prop-2-yn-1-yloxy)ethanamide, Abscisic acid receptor PYR1, MAGNESIUM ION, ... | | Authors: | Peterson, F.C, Volkman, B.F, Cutler, S.R. | | Deposit date: | 2014-11-06 | | Release date: | 2015-02-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Agrochemical control of plant water use using engineered abscisic acid receptors.
Nature, 520, 2015
|
|
4DDN
 
 | | Structure analysis of a wound-inducible lectin ipomoelin from sweet potato | | Descriptor: | Ipomoelin, methyl alpha-D-galactopyranoside | | Authors: | Chang, W.C, Liu, K.L, Jeng, S.T, Hsu, F.C, Cheng, Y.S. | | Deposit date: | 2012-01-19 | | Release date: | 2012-08-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ipomoelin, a jacalin-related lectin with a compact tetrameric association and versatile carbohydrate binding properties regulated by its N terminus.
Plos One, 7, 2012
|
|
1BVO
 
 | | DORSAL HOMOLOGUE GAMBIF1 BOUND TO DNA | | Descriptor: | DNA DUPLEX, TRANSCRIPTION FACTOR GAMBIF1 | | Authors: | Cramer, P, Varrot, A, Barillas-Mury, C, Kafatos, F.C, Mueller, C.W. | | Deposit date: | 1998-09-16 | | Release date: | 1999-07-12 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the specificity domain of the Dorsal homologue Gambif1 bound to DNA.
Structure Fold.Des., 7, 1999
|
|
5JNE
 
 | | E2-SUMO-Siz1 E3-SUMO-PCNA complex | | Descriptor: | E3 SUMO-protein ligase SIZ1,Ubiquitin-like protein SMT3, GLYCEROL, Proliferating cell nuclear antigen, ... | | Authors: | Lima, C.D, Streich Jr, F.C. | | Deposit date: | 2016-04-29 | | Release date: | 2016-08-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Capturing a substrate in an activated RING E3/E2-SUMO complex.
Nature, 536, 2016
|
|
8KHV
 
 | |
8KHW
 
 | |
