4F0Y
 
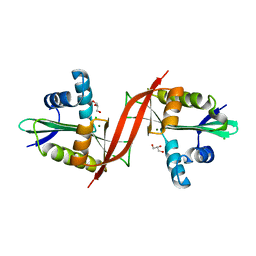 | | Crystal structure of aminoglycoside antibiotic 6'-N-acetyltransferase AAC(6')-IG from Acinetobacter haemolyticus, apo | | Descriptor: | Aminoglycoside N(6')-acetyltransferase type 1, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Evdokimova, E, Dong, A, Minasov, G, Yim, V, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-05-05 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural and Biochemical Characterization of Acinetobacter spp. Aminoglycoside Acetyltransferases Highlights Functional and Evolutionary Variation among Antibiotic Resistance Enzymes.
ACS Infect Dis., 3, 2017
|
|
3H35
 
 | | Structure of the uncharacterized protein ABO_0056 from the hydrocarbon-degrading marine bacterium Alcanivorax borkumensis SK2. | | Descriptor: | 1,2-ETHANEDIOL, S,R MESO-TARTARIC ACID, uncharacterized protein ABO_0056 | | Authors: | Cuff, M.E, Evdokimova, E, Kagan, O, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-04-15 | | Release date: | 2009-05-12 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of the uncharacterized protein ABO_0056 from the hydrocarbon-degrading marine bacterium Alcanivorax borkumensis SK2.
TO BE PUBLISHED
|
|
3HHQ
 
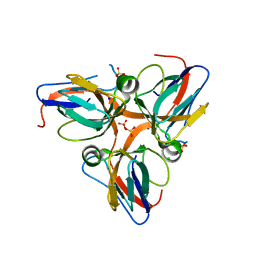 | | Crystal structure of apo dUT1p from Saccharomyces cerevisiae | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singer, A.U, Evdokimova, E, Kudritska, M, Dong, A, Edwards, A.M, Yakunin, A.F, Savchenko, A. | | Deposit date: | 2009-05-15 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and activity of the Saccharomyces cerevisiae dUTP pyrophosphatase DUT1, an essential housekeeping enzyme.
Biochem.J., 437, 2011
|
|
3HHL
 
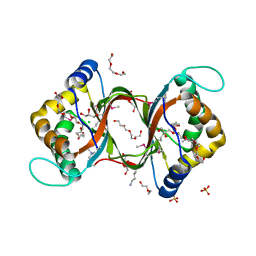 | | Crystal structure of methylated RPA0582 protein | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sledz, P, Niedzialkowska, E, Chruszcz, M, Porebski, P, Yim, V, Kudritska, M, Zimmerman, M.D, Evdokimova, E, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-15 | | Release date: | 2009-07-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of methylated RPA0582 protein
To be Published
|
|
4EBK
 
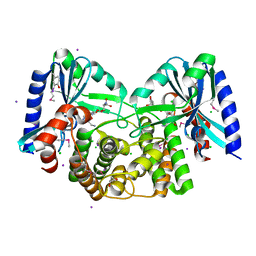 | | Crystal structure of aminoglycoside 4'-O-adenylyltransferase ANT(4')-IIb, tobramycin-bound | | Descriptor: | 1,2-ETHANEDIOL, Aminoglycoside nucleotidyltransferase, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Dong, A, Minasov, G, Evdokimova, E, Egorova, O, Yim, V, Kudritska, M, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-23 | | Release date: | 2012-04-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of aminoglycoside 4'-O-adenylyltransferase ANT(4')-IIb, tobramycin-bound
To be Published
|
|
4E8O
 
 | | Crystal structure of aminoglycoside antibiotic 6'-N-acetyltransferase AAC(6')-Ih from Acinetobacter baumannii | | Descriptor: | Aac(6')-Ih protein, CHLORIDE ION | | Authors: | Stogios, P.J, Minasov, G, Dong, A, Evdokimova, E, Yim, V, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-20 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.138 Å) | | Cite: | Structural and Biochemical Characterization of Acinetobacter spp. Aminoglycoside Acetyltransferases Highlights Functional and Evolutionary Variation among Antibiotic Resistance Enzymes.
ACS Infect Dis., 3, 2017
|
|
4ECL
 
 | | Crystal structure of the cytoplasmic domain of vancomycin resistance serine racemase VanTg | | Descriptor: | CHLORIDE ION, SULFATE ION, Serine racemase | | Authors: | Stogios, P.J, Wawrzak, Z, Minasov, G, Evdokimova, E, Egorova, O, Cosme, J, Di Leo, R, Krishnamoorthy, M, Meziane-Cherif, D, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-26 | | Release date: | 2012-04-18 | | Last modified: | 2016-06-01 | | Method: | X-RAY DIFFRACTION (2.017 Å) | | Cite: | Structural and Functional Adaptation of Vancomycin Resistance VanT Serine Racemases.
MBio, 6, 2015
|
|
4EBJ
 
 | | Crystal structure of aminoglycoside 4'-O-adenylyltransferase ANT(4')-IIb, apo | | Descriptor: | 1,2-ETHANEDIOL, Aminoglycoside nucleotidyltransferase, SULFATE ION | | Authors: | Stogios, P.J, Wawrzak, Z, Minasov, G, Evdokimova, E, Egorova, O, Yim, V, Kudritska, M, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-23 | | Release date: | 2012-04-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of aminoglycoside 4'-O-adenylyltransferase ANT(4')-IIb, apo
To be Published
|
|
3FYN
 
 | | Crystal structure from the mobile metagenome of Cole Harbour Salt Marsh: Integron Cassette Protein HFX_CASS3 | | Descriptor: | ACETATE ION, Integron gene cassette protein HFX_CASS3, MAGNESIUM ION | | Authors: | Sureshan, V, Deshpande, C.N, Harrop, S.J, Kudritska, M, Koenig, J.E, Evdokimova, E, Osipiuk, J, Edwards, A.M, Savchenko, A, Joachimiak, A, Doolittle, W.F, Stokes, H.W, Curmi, P.M.G, Mabbutt, B.C, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-01-22 | | Release date: | 2009-02-10 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | Structure from the mobile metagenome of Cole Harbour Salt Marsh: Integron Cassette Protein HFX_CASS3
To be Published
|
|
2O6T
 
 | | Crystal structure of the PA5185 protein from Pseudomonas Aeruginosa strain PAO1- orthorhombic form (P2221). | | Descriptor: | CHLORIDE ION, THIOESTERASE | | Authors: | Chruszcz, M, Koclega, K.D, Evdokimova, E, Cymborowski, M, Kudritska, M, Savchenko, A, Edwards, A, Minor, W. | | Deposit date: | 2006-12-08 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Function-biased choice of additives for optimization of protein crystallization - the case of the putative thioesterase PA5185 from Pseudomonas aeruginosa PAO1.
Cryst.Growth Des., 8, 2008
|
|
2O99
 
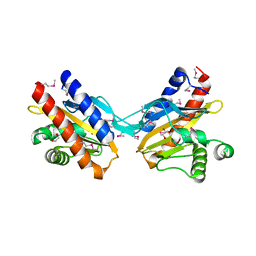 | | The crystal structure of E.coli IclR C-terminal fragment in complex with glyoxylate | | Descriptor: | 1,2-ETHANEDIOL, Acetate operon repressor, GLYCOLIC ACID | | Authors: | Lunin, V.V, Ezersky, A, Evdokimova, E, Kudritska, M, Savchenko, A. | | Deposit date: | 2006-12-13 | | Release date: | 2007-04-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Glyoxylate and Pyruvate Are Antagonistic Effectors of the Escherichia coli IclR Transcriptional Regulator.
J.Biol.Chem., 282, 2007
|
|
2O9A
 
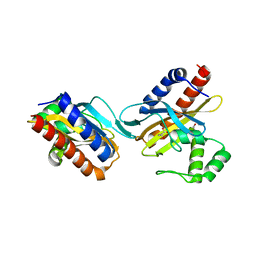 | | The crystal structure of the E.coli IclR C-terminal fragment in complex with pyruvate. | | Descriptor: | 1,2-ETHANEDIOL, Acetate operon repressor, PYRUVIC ACID | | Authors: | Lunin, V.V, Ezersky, A, Evdokimova, E, Kudritska, M, Savchenko, A. | | Deposit date: | 2006-12-13 | | Release date: | 2007-04-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Glyoxylate and Pyruvate Are Antagonistic Effectors of the Escherichia coli IclR Transcriptional Regulator.
J.Biol.Chem., 282, 2007
|
|
3GHJ
 
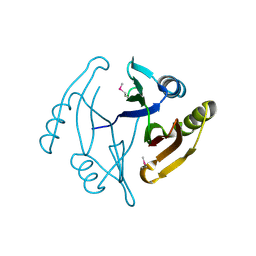 | | Crystal structure from the mobile metagenome of Halifax Harbour Sewage Outfall: Integron Cassette Protein HFX_CASS4 | | Descriptor: | Putative integron gene cassette protein | | Authors: | Sureshan, V, Deshpande, C, Harrop, S.J, Kudritska, M, Koenig, J.E, Evdokimova, E, Kim, Y, Edwards, A.M, Savchenko, A, Joachimiak, A, Doolittle, W.F, Stokes, H.W, Curmi, P.M.G, Mabbutt, B.C, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-03-03 | | Release date: | 2009-03-24 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.471 Å) | | Cite: | Structure from the mobile metagenome of Halifax Harbour Sewage Outfall: Integron Cassette Protein HFX_CASS4
To be Published
|
|
5KVK
 
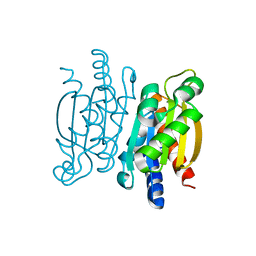 | | Crystal structure of the Competence-Damaged Protein (CinA) Superfamily Protein KP700603 from Klebsiella pneumoniae 700603 | | Descriptor: | Protein KP700603 | | Authors: | Stogios, P.J, Wawrzak, Z, Evdokimova, E, Di Leo, R, Grimshaw, S, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-14 | | Release date: | 2016-08-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | To be published
To Be Published
|
|
4EVY
 
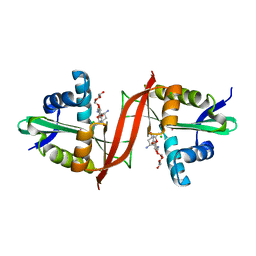 | | Crystal structure of aminoglycoside antibiotic 6'-N-acetyltransferase AAC(6')-Ig from Acinetobacter haemolyticus in complex with tobramycin | | Descriptor: | Aminoglycoside N(6')-acetyltransferase type 1, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Stogios, P.J, Evdokimova, E, Minasov, G, Yim, V, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-26 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.768 Å) | | Cite: | Structural and Biochemical Characterization of Acinetobacter spp. Aminoglycoside Acetyltransferases Highlights Functional and Evolutionary Variation among Antibiotic Resistance Enzymes.
ACS Infect Dis., 3, 2017
|
|
4F78
 
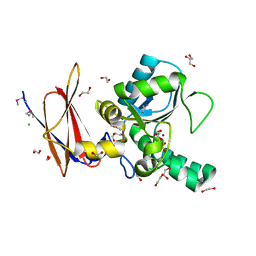 | | Crystal Structure of Vancomycin Resistance D,D-dipeptidase VanXYg | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, D,D-dipeptidase/D,D-carboxypeptidase, ... | | Authors: | Stogios, P.J, Wawrzak, Z, Evdokimova, E, Minasov, G, Egorova, O, Di Leo, R, Kudritska, M, Yim, V, Meziane-Cherif, D, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-05-15 | | Release date: | 2012-05-23 | | Last modified: | 2014-05-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for the evolution of vancomycin resistance D,D-peptidases.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4FO1
 
 | | Crystal structure of lincosamide antibiotic adenylyltransferase LnuA, apo | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Lincosamide resistance protein | | Authors: | Stogios, P.J, Wawrzak, Z, Minasov, G, Evdokimova, E, Egorova, O, Kudritska, M, Yim, V, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-06-20 | | Release date: | 2012-07-04 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of lincosamide antibiotic adenylyltransferase LnuA, apo
TO BE PUBLISHED
|
|
3CTV
 
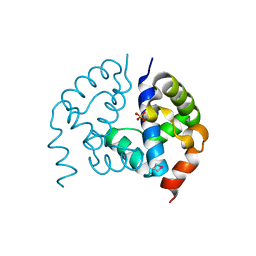 | | Crystal structure of central domain of 3-hydroxyacyl-CoA dehydrogenase from Archaeoglobus fulgidus | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, SULFATE ION | | Authors: | Osipiuk, J, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-04-14 | | Release date: | 2008-04-29 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | X-ray crystal structure of central domain of 3-hydroxyacyl-CoA dehydrogenase from Archaeoglobus fulgidus.
To be Published
|
|
2R47
 
 | | Crystal structure of MTH_862 protein of unknown function from Methanothermobacter thermautotrophicus | | Descriptor: | SULFATE ION, Uncharacterized protein MTH_862 | | Authors: | Osipiuk, J, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-08-30 | | Release date: | 2007-09-11 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure of MTH_862 protein of unknown function from Methanothermobacter thermautotrophicus.
To be Published
|
|
3ETF
 
 | | Crystal structure of a putative succinate-semialdehyde dehydrogenase from salmonella typhimurium lt2 | | Descriptor: | Putative succinate-semialdehyde dehydrogenase | | Authors: | Brunzelle, J.S, Evdokimova, E, Kudritska, M, Wawrzak, Z, Anderson, W.F, Savchenk, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-10-07 | | Release date: | 2008-11-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and activity of the NAD(P)(+) -dependent succinate semialdehyde dehydrogenase YneI from Salmonella typhimurium.
Proteins, 81, 2013
|
|
3EFV
 
 | | Crystal Structure of a Putative Succinate-Semialdehyde Dehydrogenase from Salmonella typhimurium LT2 with bound NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative succinate-semialdehyde dehydrogenase | | Authors: | Brunzelle, J.S, Evdokimova, E, Kudritska, M, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-09-10 | | Release date: | 2008-11-04 | | Last modified: | 2013-06-12 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and activity of the NAD(P)(+) -dependent succinate semialdehyde dehydrogenase YneI from Salmonella typhimurium.
Proteins, 81, 2013
|
|
4YFJ
 
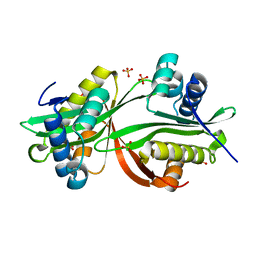 | | Crystal structure of aminoglycoside acetyltransferase AAC(3)-Ib | | Descriptor: | Aminoglycoside 3'-N-acetyltransferase, SULFATE ION | | Authors: | Stogios, P.J, Xu, Z, Evdokimova, E, Yim, V, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-02-25 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of aminoglycoside acetyltransferase AAC(3)-Ib
To Be Published
|
|
4WH5
 
 | | Crystal structure of lincosamide antibiotic adenylyltransferase LnuA, lincomycin-bound | | Descriptor: | CHLORIDE ION, LINCOMYCIN, Lincosamide resistance protein, ... | | Authors: | Stogios, P.J, Dong, A, Minasov, G, Evdokimova, E, Egorova, O, Kudritska, M, Yim, O, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-09-20 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | CRYSTAL STRUCTURE OF LINCOSAMIDE ANTIBIOTIC ADENYLYLTRANSFERASE LNUA, LINCOMYCIN BOUND
To Be Published
|
|
2PD1
 
 | | Crystal structure of NE2512 protein of unknown function from Nitrosomonas europaea | | Descriptor: | ACETATE ION, Hypothetical protein | | Authors: | Osipiuk, J, Evdokimova, E, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-03-30 | | Release date: | 2007-05-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of NE2512 protein of unknown function from Nitrosomonas europaea.
To be Published
|
|
2OMO
 
 | | Putative antibiotic biosynthesis monooxygenase from Nitrosomonas europaea | | Descriptor: | DUF176 | | Authors: | Osipiuk, J, Evdokimova, E, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-01-22 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | X-ray structure of putative antibiotic biosynthesis monooxygenase from Nitrosomonas europaea.
To be Published
|
|
