6WVX
 
 | | Crystal structure of the first bromodomain of human BRD4 with benzodiazepine inhibitor | | Descriptor: | 6'-(4-chlorophenyl)-1'-methyl-8'-(1-methyl-1H-pyrazol-4-yl)spiro[cyclopropane-1,4'-[1,2,4]triazolo[4,3-a][1,4]benzodiazepine], Bromodomain-containing protein 4 | | Authors: | Eron, S.J. | | Deposit date: | 2020-05-07 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Characterization of Degrader-Induced Ternary Complexes Using Hydrogen-Deuterium Exchange Mass Spectrometry and Computational Modeling: Implications for Structure-Based Design.
Acs Chem.Biol., 16, 2021
|
|
5K20
 
 | | Caspase-7 S239E Phosphomimetic | | Descriptor: | Caspase-7 large subunit, Caspase-7 small subunit, FORMIC ACID | | Authors: | Eron, S.J, Hardy, J.A. | | Deposit date: | 2016-05-18 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | PAK2 Phosphorylation Inhibits Caspase-7 by Two Divergent Mechanisms: Slowing Activation and Blocking Substrate Binding
To Be Published
|
|
7N03
 
 | | Crystal structure of MTH1 in complex with compound 31 | | Descriptor: | 4-anilino-6-(hexylamino)-N-methylquinoline-3-carboxamide, 7,8-dihydro-8-oxoguanine triphosphatase, SULFATE ION | | Authors: | Eron, S.J. | | Deposit date: | 2021-05-24 | | Release date: | 2021-11-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Development of an AchillesTAG degradation system and its application to control CAR-T activity
Curr Res Chem Biol, 1, 2021
|
|
7N13
 
 | | Crystal structure of MTH1 in complex with compound 32 | | Descriptor: | 4-anilino-6-[4-(butylcarbamoyl)-3-fluorophenyl]-N-cyclopropyl-7-fluoroquinoline-3-carboxamide, 7,8-dihydro-8-oxoguanine triphosphatase, SULFATE ION | | Authors: | Eron, S.J. | | Deposit date: | 2021-05-26 | | Release date: | 2021-11-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Development of an AchillesTAG degradation system and its application to control CAR-T activity
Curr Res Chem Biol, 1, 2021
|
|
6ZEI
 
 | | Structure of PP1-IRSp53 S455E chimera [PP1(7-304) + linker (G/S)x9 + IRSp53(449-465)] bound to Phactr1 (516-580) | | Descriptor: | GLYCEROL, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Mouilleron, S, Treisman, R, Fedoryshchak, R, Lee, R, Butler, A.M, Prechova, M. | | Deposit date: | 2020-06-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Molecular basis for substrate specificity of the Phactr1/PP1 phosphatase holoenzyme.
Elife, 9, 2020
|
|
6ZEH
 
 | | Structure of PP1-spectrin alpha II chimera [PP1(7-304) + linker (G/S)x9 + spectrin alpha II (1025-1039)] bound to Phactr1 (516-580) | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, Phosphatase and actin regulator, ... | | Authors: | Mouilleron, S, Treisman, R, Fedoryshchak, R, Lee, R, Butler, A.M, Prechova, M. | | Deposit date: | 2020-06-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular basis for substrate specificity of the Phactr1/PP1 phosphatase holoenzyme.
Elife, 9, 2020
|
|
6ZEE
 
 | | Structure of PP1(7-300) bound to Phactr1 (507-580) at pH8.4 | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE, GLYCEROL, ... | | Authors: | Mouilleron, S, Treisman, R, Fedoryshchak, R, Lee, R, Butler, A.M, Prechova, M. | | Deposit date: | 2020-06-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis for substrate specificity of the Phactr1/PP1 phosphatase holoenzyme.
Elife, 9, 2020
|
|
6ZEF
 
 | | Structure of PP1(7-300) bound to Phactr1 (516-580) at pH 5.25 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Mouilleron, S, Treisman, R, Fedoryshchak, R, Lee, R, Butler, A.M, Prechova, M. | | Deposit date: | 2020-06-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Molecular basis for substrate specificity of the Phactr1/PP1 phosphatase holoenzyme.
Elife, 9, 2020
|
|
6ZEG
 
 | | Structure of PP1-IRSp53 chimera [PP1(7-304) + linker (G/S)x9 + IRSp53(449-465)] bound to Phactr1 (516-580) | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE, MANGANESE (II) ION, ... | | Authors: | Mouilleron, S, Treisman, R, Fedoryshchak, R, Lee, R, Butler, A.M, Prechova, M. | | Deposit date: | 2020-06-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Molecular basis for substrate specificity of the Phactr1/PP1 phosphatase holoenzyme.
Elife, 9, 2020
|
|
2YJF
 
 | | Oligomeric assembly of actin bound to MRTF-A | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Mouilleron, S, Langer, C.A, Guettler, S, McDonald, N.Q, Treisman, R. | | Deposit date: | 2011-05-19 | | Release date: | 2011-07-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of a pentavalent G-actin*MRTF-A complex reveals how G-actin controls nucleocytoplasmic shuttling of a transcriptional coactivator.
Sci Signal, 4, 2011
|
|
4UC0
 
 | | Crystal Structure Of a purine nucleoside phosphorylase (PSI-NYSGRC-029736) from Agrobacterium vitis | | Descriptor: | HYPOXANTHINE, Purine nucleoside phosphorylase | | Authors: | Cameron, S.A, Sampathkumar, P, Ramagopal, U.A, Attonito, J, Ahmed, M, Bhosle, R, Bonanno, J, Chamala, S, Chowdhury, S, Glenn, A.S, Hammonds, J, Hillerich, B, Love, J.D, Seidel, R, Stead, M, Toro, R, Wasserman, S.R, Schramm, V.L, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-08-13 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure Of a purine nucleoside phosphorylase (PSI-NYSGRC-029736) from Agrobacterium vitis
To be published
|
|
5LXH
 
 | | GABARAP-L1 ATG4B LIR Complex | | Descriptor: | Cysteine protease ATG4B, GLYCEROL, Gamma-aminobutyric acid receptor-associated protein-like 1, ... | | Authors: | Mouilleron, S, Skytte Rasmussen, M, Kumar Shrestha, B, Wirth, M, Bowitz Larsen, K, Abudu Princely, Y, Sjottem, E, Tooze, S, Lamark, T, Johansen, T, Lee, R. | | Deposit date: | 2016-09-21 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | ATG4B contains a C-terminal LIR motif important for binding and efficient cleavage of mammalian orthologs of yeast Atg8.
Autophagy, 13, 2017
|
|
5LXI
 
 | | GABARAP-L1 ATG4B LIR Complex | | Descriptor: | 1,2-ETHANEDIOL, Cysteine protease ATG4B, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mouilleron, S, Skytte Rasmussen, M, Kumar Shrestha, B, Wirth, M, Bowitz Larsen, K, Abudu Princely, Y, Sjottem, E, Tooze, S, Lamark, T, Johansen, T, Lee, R. | | Deposit date: | 2016-09-21 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | ATG4B contains a C-terminal LIR motif important for binding and efficient cleavage of mammalian orthologs of yeast Atg8.
Autophagy, 13, 2017
|
|
6ZEJ
 
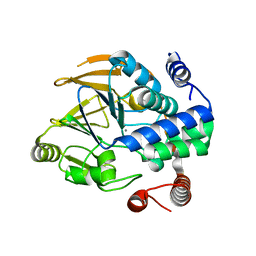 | | Structure of PP1-Phactr1 chimera [PP1(7-304) + linker (SGSGS) + Phactr1(526-580)] | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Mouilleron, S, Treisman, R, Fedoryshchak, R, Lee, R, Butler, A.M, Prechova, M. | | Deposit date: | 2020-06-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Molecular basis for substrate specificity of the Phactr1/PP1 phosphatase holoenzyme.
Elife, 9, 2020
|
|
6YOO
 
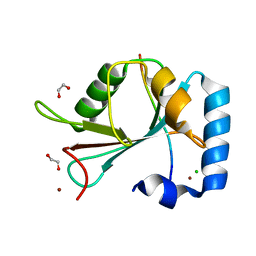 | | Structure of SAMM50 LIR bound to GABARAPL1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Gamma-aminobutyric acid receptor-associated protein-like 1, ... | | Authors: | Mouilleron, S, Wenxin, Z, Johansen, T, Tooze, S, Abudu, Y.P, Lamark, T. | | Deposit date: | 2020-04-14 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Structure of SAMM50 LIR bound to GABARAPL1
To Be Published
|
|
6YOP
 
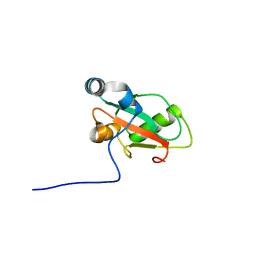 | | Structure of SAMM50 LIR bound to GABARAP | | Descriptor: | Sorting and assembly machinery component 50 homolog,Gamma-aminobutyric acid receptor-associated protein | | Authors: | Mouilleron, S, Wenxin, Z, Johansen, T, Tooze, S, Abudu, Y.P, Lamark, T. | | Deposit date: | 2020-04-14 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure of SAMM50 LIR bound to GABARAP
To Be Published
|
|
5TC6
 
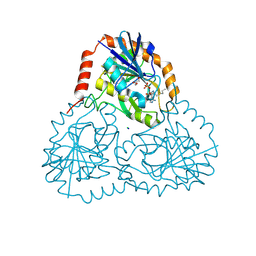 | | Crystal structure of human 5'-deoxy-5'-methylthioadenosine phosphorylase in complex with propylthio-immucillin-A | | Descriptor: | (2S,3S,4R,5S)-2-(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)-5-[(propylsulfanyl)methyl]pyrrolidine-3,4-diol, CHLORIDE ION, GLYCEROL, ... | | Authors: | Cameron, S.A, Firestone, R.S, Schramm, V.L, Almo, S.C. | | Deposit date: | 2016-09-14 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | TBA
To be published
|
|
5TC7
 
 | | Crystal structure of human 5'-deoxy-5'-methylthioadenosine phosphorylase in complex with 5'-methylthiotubercidin at 1.75 angstrom | | Descriptor: | 2-(4-AMINO-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-METHYLSULFANYLMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Cameron, S.A, Firestone, R.S, Schramm, V.L, Almo, S.C. | | Deposit date: | 2016-09-14 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | TBA
To be published
|
|
5TC8
 
 | | Crystal structure of human 5'-deoxy-5'-methylthioadenosine phosphorylase in complex with methylthio-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)METHYL]-4-[(METHYLSULFANYL)METHYL]PYRROLIDIN-3-OL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Cameron, S.A, Firestone, R.S, Schramm, V.L, Almo, S.C. | | Deposit date: | 2016-09-14 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | TBA
To be published
|
|
5TC5
 
 | | Crystal structure of human 5'-deoxy-5'-methylthioadenosine phosphorylase in complex with butylthio-DADMe-Immucillin-A and chloride | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-[(butylsulfanyl)methyl]pyrrolidin-3-ol, CHLORIDE ION, S-methyl-5'-thioadenosine phosphorylase | | Authors: | Cameron, S.A, Firestone, R.S, Schramm, V.L, Almo, S.C. | | Deposit date: | 2016-09-14 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | TBA
To be published
|
|
4WKC
 
 | | Crystal structure of Escherichia coli 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with butylthio-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-[(butylsulfanyl)methyl]pyrrolidin-3-ol, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, TETRAETHYLENE GLYCOL | | Authors: | Cameron, S.A, Thomas, K, Almo, S.C, Schramm, V.L. | | Deposit date: | 2014-10-02 | | Release date: | 2015-08-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Active site and remote contributions to catalysis in methylthioadenosine nucleosidases.
Biochemistry, 54, 2015
|
|
5EUB
 
 | | Crystal structure of human 5'-deoxy-5'-methylthioadenosine phosphorylase in complex with 2-amino-MTA and sulfate | | Descriptor: | (2~{R},3~{R},4~{S},5~{S})-2-[2,6-bis(azanyl)purin-9-yl]-5-(methylsulfanylmethyl)oxolane-3,4-diol, CHLORIDE ION, GLYCEROL, ... | | Authors: | Cameron, S.A, Firestone, R.S, Schramm, V.L, Almo, S.C. | | Deposit date: | 2015-11-18 | | Release date: | 2016-12-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structure of human 5'-deoxy-5'-methylthioadenosine phosphorylase in complex with 2-amino-MTA and sulfate
To be published
|
|
4WKB
 
 | | Crystal structure of Vibrio cholerae 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with methylthio-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)METHYL]-4-[(METHYLSULFANYL)METHYL]PYRROLIDIN-3-OL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase | | Authors: | Cameron, S.A, Thomas, K, Almo, S.C, Schramm, V.L. | | Deposit date: | 2014-10-02 | | Release date: | 2015-08-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Active site and remote contributions to catalysis in methylthioadenosine nucleosidases.
Biochemistry, 54, 2015
|
|
4WKN
 
 | | Crystal structure of Helicobacter pylori 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with methylthio-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)METHYL]-4-[(METHYLSULFANYL)METHYL]PYRROLIDIN-3-OL, Aminodeoxyfutalosine nucleosidase | | Authors: | Cameron, S.A, Wang, S, Almo, S.C, Schramm, V.L. | | Deposit date: | 2014-10-02 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | New Antibiotic Candidates against Helicobacter pylori.
J.Am.Chem.Soc., 137, 2015
|
|
4WKO
 
 | | Crystal structure of Helicobacter pylori 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with hydroxybutylthio-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-{[(4-hydroxybutyl)sulfanyl]methyl}pyrrolidin-3-ol, Aminodeoxyfutalosine nucleosidase | | Authors: | Cameron, S.A, Wang, S, Almo, S.C, Schramm, V.L. | | Deposit date: | 2014-10-02 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | New Antibiotic Candidates against Helicobacter pylori.
J.Am.Chem.Soc., 137, 2015
|
|
