4YNB
 
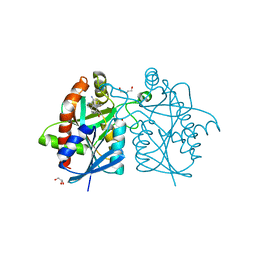 | | Crystal structure of Helicobacter pylori 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with pyrazinylthio-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-[(pyrazin-2-ylsulfanyl)methyl]pyrrolidin-3-ol, Aminodeoxyfutalosine nucleosidase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Cameron, S.A, Wang, S, Almo, S.C, Schramm, V.L. | | Deposit date: | 2015-03-09 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | New Antibiotic Candidates against Helicobacter pylori.
J.Am.Chem.Soc., 137, 2015
|
|
4YO8
 
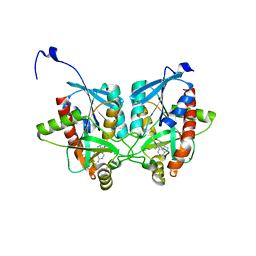 | | Crystal structure of Helicobacter pylori 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with (((4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl)(hexyl)amino)methanol | | Descriptor: | Aminodeoxyfutalosine nucleosidase, ZINC ION, {[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl](hexyl)amino}methanol | | Authors: | Cameron, S.A, Wang, S, Almo, S.C, Schramm, V.L. | | Deposit date: | 2015-03-11 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New Antibiotic Candidates against Helicobacter pylori.
J.Am.Chem.Soc., 137, 2015
|
|
4WKP
 
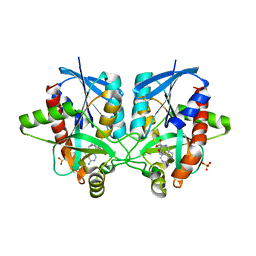 | | Crystal structure of Helicobacter pylori 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with 2-(2-hydroxyethoxy)ethylthiomethyl-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-(2-{[2-(2-hydroxyethoxy)ethyl]sulfanyl}ethyl)pyrrolidin-3-ol, Aminodeoxyfutalosine nucleosidase, SULFATE ION | | Authors: | Cameron, S.A, Wang, S, Almo, S.C, Schramm, V.L. | | Deposit date: | 2014-10-02 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | New Antibiotic Candidates against Helicobacter pylori.
J.Am.Chem.Soc., 137, 2015
|
|
4YML
 
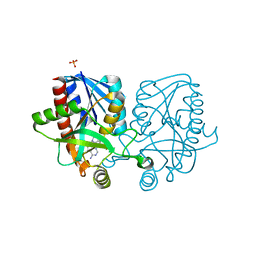 | | Crystal structure of Escherichia coli 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with (3S,4R)-methylthio-DADMe-Immucillin-A | | Descriptor: | (3S,4R)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-[(methylsulfanyl)methyl]pyrrolidin-3-ol, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, PHOSPHATE ION | | Authors: | Cameron, S.A, Thomas, K, Almo, S.C, Schramm, V.L. | | Deposit date: | 2015-03-06 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Tight binding enantiomers of pre-clinical drug candidates.
Bioorg.Med.Chem., 23, 2015
|
|
4WKN
 
 | | Crystal structure of Helicobacter pylori 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with methylthio-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)METHYL]-4-[(METHYLSULFANYL)METHYL]PYRROLIDIN-3-OL, Aminodeoxyfutalosine nucleosidase | | Authors: | Cameron, S.A, Wang, S, Almo, S.C, Schramm, V.L. | | Deposit date: | 2014-10-02 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | New Antibiotic Candidates against Helicobacter pylori.
J.Am.Chem.Soc., 137, 2015
|
|
4WKO
 
 | | Crystal structure of Helicobacter pylori 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with hydroxybutylthio-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-{[(4-hydroxybutyl)sulfanyl]methyl}pyrrolidin-3-ol, Aminodeoxyfutalosine nucleosidase | | Authors: | Cameron, S.A, Wang, S, Almo, S.C, Schramm, V.L. | | Deposit date: | 2014-10-02 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | New Antibiotic Candidates against Helicobacter pylori.
J.Am.Chem.Soc., 137, 2015
|
|
6GHM
 
 | | Structure of PP1 alpha phosphatase bound to ASPP2 | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Apoptosis-stimulating of p53 protein 2, ... | | Authors: | Mouilleron, S, Bertran, T.M, Tapon, N, Zhou, Y. | | Deposit date: | 2018-05-08 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | ASPP proteins discriminate between PP1 catalytic subunits through their SH3 domain and the PP1 C-tail.
Nat Commun, 10, 2019
|
|
6GVC
 
 | | Structure of ArhGAP12 bound to G-Actin | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Mouilleron, S, Treisman, R, Diring, J. | | Deposit date: | 2018-06-20 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | RPEL-family rhoGAPs link Rac/Cdc42 GTP loading to G-actin availability.
Nat.Cell Biol., 21, 2019
|
|
2YJE
 
 | | Oligomeric assembly of actin bound to MRTF-A | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Mouilleron, S, Langer, C.A, Guettler, S, McDonald, N.Q, Treisman, R. | | Deposit date: | 2011-05-19 | | Release date: | 2011-07-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of a pentavalent G-actin*MRTF-A complex reveals how G-actin controls nucleocytoplasmic shuttling of a transcriptional coactivator.
Sci Signal, 4, 2011
|
|
6G86
 
 | | Structure of Cdc14 bound to SIC1 PxL motif | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, HEXAETHYLENE GLYCOL, ... | | Authors: | Mouilleron, S, Kataria, M, Uhlmann, F. | | Deposit date: | 2018-04-07 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | A PxL motif promotes timely cell cycle substrate dephosphorylation by the Cdc14 phosphatase.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6HYL
 
 | | Structure of PCM1 LIR motif bound to GABARAP | | Descriptor: | Pericentriolar material 1 protein,Gamma-aminobutyric acid receptor-associated protein | | Authors: | Mouilleron, S, Wirth, M, Zhang, W, O'Reilly, N, Tooze, S, Johansen, T, Razi, M, Nyoni, L, Joshi, D. | | Deposit date: | 2018-10-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.559 Å) | | Cite: | Molecular determinants regulating selective binding of autophagy adapters and receptors to ATG8 proteins.
Nat Commun, 10, 2019
|
|
6HOG
 
 | | Structure of VPS34 LIR motif bound to GABARAP | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Phosphatidylinositol 3-kinase catalytic subunit type 3,Gamma-aminobutyric acid receptor-associated protein, ... | | Authors: | Mouilleron, S, Birgisdottir, A.B, Bhujbal, Z, Wirth, M, Sjottem, E, Evjen, G, Zhang, W, Lee, R, O'Reilly, N, Tooze, S, Lamark, T, Johansen, T. | | Deposit date: | 2018-09-17 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Members of the autophagy class III phosphatidylinositol 3-kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs.
Autophagy, 15, 2019
|
|
6HOK
 
 | | Structure of Beclin1 LIR (S96E) motif bound to GABARAP | | Descriptor: | 1,2-ETHANEDIOL, Beclin-1,Gamma-aminobutyric acid receptor-associated protein | | Authors: | Mouilleron, S, Birgisdottir, A.B, Bhujbal, Z, Wirth, M, Sjottem, E, Evjen, G, Zhang, W, Lee, R, O'Reilly, N, Tooze, S, Lamark, T, Johansen, T. | | Deposit date: | 2018-09-17 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Members of the autophagy class III phosphatidylinositol 3-kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs.
Autophagy, 15, 2019
|
|
6HOI
 
 | | Structure of Beclin1 LIR motif bound to GABARAPL1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Beclin-1, ... | | Authors: | Mouilleron, S, Birgisdottir, A.B, Bhujbal, Z, Wirth, M, Sjottem, E, Evjen, G, Zhang, W, Lee, R, O'Reilly, N, Tooze, S, Lamark, T, Johansen, T. | | Deposit date: | 2018-09-17 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Members of the autophagy class III phosphatidylinositol 3-kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs.
Autophagy, 15, 2019
|
|
6HYM
 
 | | Structure of PCM1 LIR motif bound to GABARAP | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Pericentriolar material 1 protein,Gamma-aminobutyric acid receptor-associated protein | | Authors: | Mouilleron, S, Wirth, M, Zhang, W, O'Reilly, N, Tooze, S, Johansen, T, Razi, M, Nyoni, L, Joshi, D. | | Deposit date: | 2018-10-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Molecular determinants regulating selective binding of autophagy adapters and receptors to ATG8 proteins.
Nat Commun, 10, 2019
|
|
6HOH
 
 | | Structure of VPS34 LIR motif (S249E) bound to GABARAP | | Descriptor: | Phosphatidylinositol 3-kinase catalytic subunit type 3,Gamma-aminobutyric acid receptor-associated protein, TRIETHYLENE GLYCOL | | Authors: | Mouilleron, S, Birgisdottir, A.B, Bhujbal, Z, Wirth, M, Sjottem, E, Evjen, G, Zhang, W, Lee, R, O'Reilly, N, Tooze, S, Lamark, T, Johansen, T. | | Deposit date: | 2018-09-17 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Members of the autophagy class III phosphatidylinositol 3-kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs.
Autophagy, 15, 2019
|
|
6G84
 
 | | Structure of Cdc14 bound to CBK1 PxL motif | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CBK1, ... | | Authors: | Mouilleron, S, Kataria, M, Uhlmann, F. | | Deposit date: | 2018-04-07 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | A PxL motif promotes timely cell cycle substrate dephosphorylation by the Cdc14 phosphatase.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6G85
 
 | | Structure of Cdc14 bound to CBK1 PxL motif | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CBK1, ... | | Authors: | Mouilleron, S, Kataria, M, Uhlmann, F. | | Deposit date: | 2018-04-07 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.528 Å) | | Cite: | A PxL motif promotes timely cell cycle substrate dephosphorylation by the Cdc14 phosphatase.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6HYN
 
 | | Structure of ATG13 LIR motif bound to GABARAP | | Descriptor: | Autophagy-related protein 13,Gamma-aminobutyric acid receptor-associated protein | | Authors: | Mouilleron, S, Wirth, M, Zhang, W, O'Reilly, N, Tooze, S, Johansen, T, Razi, M, Nyoni, L, Joshi, D. | | Deposit date: | 2018-10-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Molecular determinants regulating selective binding of autophagy adapters and receptors to ATG8 proteins.
Nat Commun, 10, 2019
|
|
6HOL
 
 | | Structure of ATG14 LIR motif bound to GABARAPL1 | | Descriptor: | Beclin 1-associated autophagy-related key regulator, GLYCEROL, Gamma-aminobutyric acid receptor-associated protein-like 1, ... | | Authors: | Mouilleron, S, Birgisdottir, A.B, Bhujbal, Z, Wirth, M, Sjottem, E, Evjen, G, Zhang, W, Lee, R, O'Reilly, N, Tooze, S, Lamark, T, Johansen, T. | | Deposit date: | 2018-09-17 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Members of the autophagy class III phosphatidylinositol 3-kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs.
Autophagy, 15, 2019
|
|
6HOJ
 
 | | Structure of Beclin1 LIR motif bound to GABARAP | | Descriptor: | 1,2-ETHANEDIOL, Beclin-1,Gamma-aminobutyric acid receptor-associated protein, SULFATE ION | | Authors: | Mouilleron, S, Birgisdottir, A.B, Bhujbal, Z, Wirth, M, Sjottem, E, Evjen, G, Zhang, W, Lee, R, O'Reilly, N, Tooze, S, Lamark, T, Johansen, T. | | Deposit date: | 2018-09-17 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Members of the autophagy class III phosphatidylinositol 3-kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs.
Autophagy, 15, 2019
|
|
6HYO
 
 | | Structure of ULK1 LIR motif bound to GABARAP | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Mouilleron, S, Wirth, M, Zhang, W, O'Reilly, N, Tooze, S, Johansen, T, Razi, M, Nyoni, L, Joshi, D. | | Deposit date: | 2018-10-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Molecular determinants regulating selective binding of autophagy adapters and receptors to ATG8 proteins.
Nat Commun, 10, 2019
|
|
2V52
 
 | | Structure of MAL-RPEL2 complexed to G-actin | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Mouilleron, S, Guettler, S, Langer, C.A, Treisman, R, McDonald, N.Q. | | Deposit date: | 2008-10-01 | | Release date: | 2008-11-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Molecular basis for G-actin binding to RPEL motifs from the serum response factor coactivator MAL.
EMBO J., 27, 2008
|
|
2V6U
 
 | |
3OOJ
 
 | |
