1TK8
 
 | | T7 DNA polymerase ternary complex with 8 oxo guanosine and dAMP at the elongation site | | Descriptor: | 2',3'-DIDEOXY-THYMIDINE-5'-TRIPHOSPHATE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5'-D(*CP*CP*CP*AP*(8OG)P*TP*GP*GP*CP*AP*CP*TP*GP*GP*CP*CP*GP*TP*CP*GP*TP*TP*TP*TP*CP*G)-3', ... | | Authors: | Brieba, L.G, Eichman, B.F, Kokoska, R.J, Doublie, S, Kunkel, T.A, Ellenberger, T. | | Deposit date: | 2004-06-08 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the dual coding potential of 8-oxoguanosine by a high-fidelity DNA polymerase.
Embo J., 23, 2004
|
|
5UUG
 
 | |
5VI0
 
 | |
5VHV
 
 | | Pseudomonas fluorescens alkylpurine DNA glycosylase AlkC bound to DNA containing an oxocarbenium-intermediate analog | | Descriptor: | (2R,5R,13R,16R)-9-(hydroxymethyl)-9-{[(2R)-2-hydroxypropoxy]methyl}-5,13-dimethyl-4,7,11,14-tetraoxaheptadecane-2,16-diol, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA (5'-D(*AP*AP*GP*AP*CP*TP*TP*GP*GP*AP*C)-3'), ... | | Authors: | Shi, R, Eichman, B.F. | | Deposit date: | 2017-04-13 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Selective base excision repair of DNA damage by the non-base-flipping DNA glycosylase AlkC.
EMBO J., 37, 2018
|
|
2OFI
 
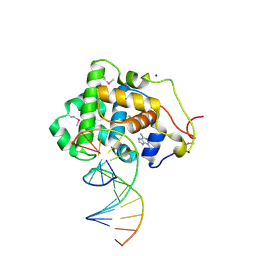 | | Crystal Structure of 3-methyladenine DNA Glycosylase I (TAG) bound to DNA/3mA | | Descriptor: | 3-METHYL-3H-PURIN-6-YLAMINE, 3-methyladenine DNA glycosylase I, constitutive, ... | | Authors: | Metz, A.H, Hollis, T, Eichman, B.F. | | Deposit date: | 2007-01-03 | | Release date: | 2007-05-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | DNA damage recognition and repair by 3-methyladenine DNA glycosylase I (TAG).
Embo J., 26, 2007
|
|
5UUJ
 
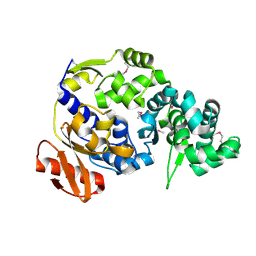 | |
1TK5
 
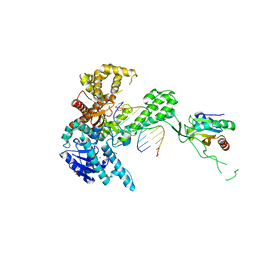 | | T7 DNA polymerase binary complex with 8 oxo guanosine in the templating strand | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5'-D(*CP*CP*CP*(8OG)P*CP*TP*GP*GP*CP*AP*CP*TP*GP*GP*CP*CP*GP*TP*CP*GP*TP*TP*TP*TP*CP*G)-3', 5'-D(*CP*GP*AP*AP*A*GP*CP*CP*AP*GP*TP*GP*CP*CP*AP*(DDG)P*TP*GP*CP*AP*A)-3', ... | | Authors: | Brieba, L.G, Eichman, B.F, Kokoska, R.J, Doublie, S, Kunkel, T.A, Ellenberger, T. | | Deposit date: | 2004-06-08 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the dual coding potential of 8-oxoguanosine by a high-fidelity DNA polymerase.
Embo J., 23, 2004
|
|
1TK0
 
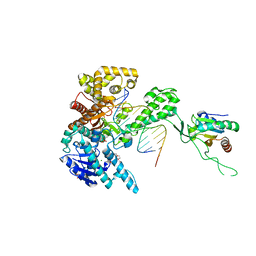 | | T7 DNA polymerase ternary complex with 8 oxo guanosine and ddCTP at the insertion site | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5'-D(*CP*CP*CP*(8OG)P*CP*TP*GP*GP*CP*AP*CP*TP*GP*GP*CP*CP*GP*TP*CP*GP*TP*TP*TP*TP*CP*G)-3', ... | | Authors: | Brieba, L.G, Eichman, B.F, Kokoska, R.J, Doublie, S, Kunkel, T.A, Ellenberger, T. | | Deposit date: | 2004-06-07 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the dual coding potential of 8-oxoguanosine by a high-fidelity DNA polymerase.
Embo J., 23, 2004
|
|
2OFK
 
 | | Crystal Structure of 3-methyladenine DNA glycosylase I (TAG) | | Descriptor: | 3-methyladenine DNA glycosylase I, constitutive, TRIETHYLENE GLYCOL, ... | | Authors: | Metz, A.H, Hollis, T, Eichman, B.F. | | Deposit date: | 2007-01-03 | | Release date: | 2007-05-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | DNA damage recognition and repair by 3-methyladenine DNA glycosylase I (TAG).
Embo J., 26, 2007
|
|
5UUF
 
 | |
5UUH
 
 | |
1T8E
 
 | | T7 DNA Polymerase Ternary Complex with dCTP at the Insertion Site. | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 25-MER, ... | | Authors: | Brieba, L.G, Eichman, B.F, Kokoska, R.J, Doublie, S, Kunkel, T.A, Ellenberger, T. | | Deposit date: | 2004-05-12 | | Release date: | 2004-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structural basis for the dual coding potential of 8-oxoguanosine by a high-fidelity DNA polymerase.
Embo J., 23, 2004
|
|
1TKD
 
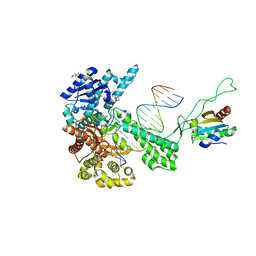 | | T7 DNA polymerase ternary complex with 8 oxo guanosine and dCMP at the elongation site | | Descriptor: | 2',3'-DIDEOXY-THYMIDINE-5'-TRIPHOSPHATE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA (5'-D(*CP*CP*CP*AP*(8OG)P*TP*GP*GP*CP*AP*CP*TP*GP*GP*CP*CP*GP*TP*CP*GP*TP*TP*TP*TP*CP*G)-3'), ... | | Authors: | Brieba, L.G, Eichman, B.F, Kokoska, R.J, Doublie, S, Kunkel, T.A, Ellenberger, T. | | Deposit date: | 2004-06-08 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural basis for the dual coding potential of 8-oxoguanosine by a high-fidelity DNA polymerase.
Embo J., 23, 2004
|
|
2KWQ
 
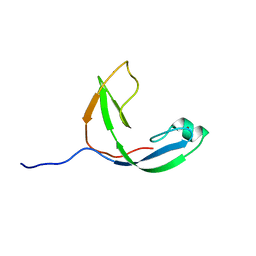 | | Mcm10 C-terminal DNA binding domain | | Descriptor: | Protein MCM10 homolog, ZINC ION | | Authors: | Robertson, P.D, Chagot, B, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2010-04-15 | | Release date: | 2010-05-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the C-terminal DNA binding domain of Mcm10 reveals a conserved MCM motif.
J.Biol.Chem., 285, 2010
|
|
1F69
 
 | |
1F6C
 
 | |
1F6E
 
 | |
1F6I
 
 | |
1F6J
 
 | |
5CLB
 
 | |
5CL3
 
 | |
5CL8
 
 | |
5CLE
 
 | |
5CL9
 
 | |
5CL4
 
 | |
