8G9N
 
 | | Partial DNA elongation subcomplex of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA polymerase alpha catalytic subunit, DNA template, ... | | Authors: | Mullins, E.A, Durie, C.L, Ohi, M.D, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-02-21 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8G99
 
 | | Partial auto-inhibitory complex of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, DNA primase large subunit, ... | | Authors: | Mullins, E.A, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-02-21 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7LXJ
 
 | |
7LXH
 
 | | Bacillus cereus DNA glycosylase AlkD bound to a CC1065-adenine nucleobase adduct and DNA containing an abasic site | | Descriptor: | 7-{7-[(1R)-1-{[(4P)-6-amino-3H-purin-3-yl]methyl}-5-hydroxy-8-methyl-1,6-dihydropyrrolo[3,2-e]indole-3(2H)-carbonyl]-4-hydroxy-5-methoxy-1,6-dihydropyrrolo[3,2-e]indole-3(2H)-carbonyl}-4-hydroxy-5-methoxy-1,6-dihydropyrrolo[3,2-e]indole-3(2H)-carboxamide, CALCIUM ION, DNA (5'-D(*AP*GP*CP*AP*AP*(ORP)P*GP*GP*C)-3'), ... | | Authors: | Mullins, E.A, Eichman, B.F. | | Deposit date: | 2021-03-03 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.667 Å) | | Cite: | Structural evolution of a DNA repair self-resistance mechanism targeting genotoxic secondary metabolites.
Nat Commun, 12, 2021
|
|
3EBE
 
 | |
8UCU
 
 | | Partial DNA termination subcomplex of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA polymerase alpha catalytic subunit, DNA template, ... | | Authors: | Mullins, E.A, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-09-27 | | Release date: | 2023-10-11 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8UCW
 
 | | Complete DNA termination subcomplex 2 of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA polymerase alpha catalytic subunit, DNA template, ... | | Authors: | Mullins, E.A, Chazin, W.C, Eichman, B.F. | | Deposit date: | 2023-09-27 | | Release date: | 2023-10-11 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8UCV
 
 | | Complete DNA termination subcomplex 1 of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA polymerase alpha catalytic subunit, DNA primase large subunit, ... | | Authors: | Mullins, E.A, Chazin, W.C, Eichman, B.F. | | Deposit date: | 2023-09-27 | | Release date: | 2023-10-11 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8V6J
 
 | | DNA elongation complex (configuration 2) of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, ... | | Authors: | Mullins, E.A, Durie, C.L, Ohi, M.D, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-12-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (11.11 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8V5O
 
 | | Tetramer core subcomplex (conformation 3) of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, DNA primase, ... | | Authors: | Mullins, E.A, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-11-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (8.99 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8V5N
 
 | | Tetramer core subcomplex (conformation 2) of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, DNA primase, ... | | Authors: | Mullins, E.A, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-11-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (8.56 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8V6I
 
 | | DNA elongation complex (configuration 1) of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, ... | | Authors: | Mullins, E.A, Durie, C.L, Ohi, M.D, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-12-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (14.06 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8V6G
 
 | | DNA initiation complex (configuration 1) of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, ... | | Authors: | Mullins, E.A, Durie, C.L, Ohi, M.D, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-12-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (11.16 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8V5M
 
 | | Tetramer core subcomplex (conformation 1) of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, DNA primase, ... | | Authors: | Mullins, E.A, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-11-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (9.22 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8V6H
 
 | | DNA initiation complex (configuration 2) of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, ... | | Authors: | Mullins, E.A, Durie, C.L, Ohi, M.D, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-12-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (11.11 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
4LIK
 
 | |
4JBZ
 
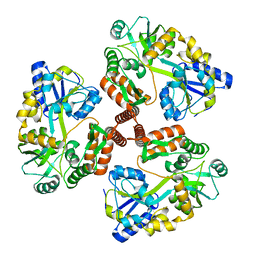 | | Structure of Mcm10 coiled-coil region | | Descriptor: | MALTOSE-BINDING PERIPLASMIC PROTEIN FUSED WITH XENOPUS LAEVIS MCM10 COILED-COIL REGION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Du, W, Adhikary, S, Eichman, B.F. | | Deposit date: | 2013-02-20 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mcm10 self-association is mediated by an N-terminal coiled-coil domain.
Plos One, 8, 2013
|
|
4LIL
 
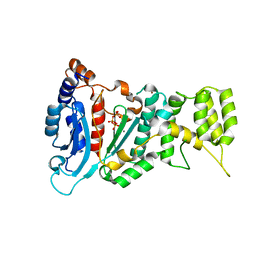 | | Crystal structure of the catalytic subunit of human primase bound to UTP and Mn | | Descriptor: | DNA primase small subunit, MANGANESE (II) ION, URIDINE 5'-TRIPHOSPHATE, ... | | Authors: | Vaithiyalingam, S, Eichman, B.F, Chazin, W.J. | | Deposit date: | 2013-07-02 | | Release date: | 2013-12-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insights into Eukaryotic Primer Synthesis from Structures of the p48 Subunit of Human DNA Primase.
J.Mol.Biol., 426, 2014
|
|
3H15
 
 | |
3JXY
 
 | |
3JXZ
 
 | |
3JY1
 
 | |
3JX7
 
 | |
4OU0
 
 | |
4O66
 
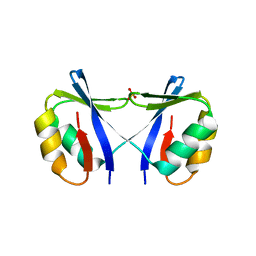 | | Crystal Structure of SMARCAL1 HARP substrate recognition domain | | Descriptor: | SODIUM ION, SULFATE ION, SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 | | Authors: | Mason, A.C, Eichman, B.F. | | Deposit date: | 2013-12-20 | | Release date: | 2014-05-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A structure-specific nucleic acid-binding domain conserved among DNA repair proteins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
