1TPE
 
 | |
2V5L
 
 | | Structures of the Open and Closed State of Trypanosomal Triosephosphate Isomerase: as Observed in a New Crystal Form: Implications for the Reaction Mechanism | | Descriptor: | SULFATE ION, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Noble, M.E.M, Zeelen, J.P, Wierenga, R.K. | | Deposit date: | 2007-07-06 | | Release date: | 2007-07-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of the Open and Closed State of Trypanosomal Triosephosphate Isomerase: As Observed in a New Crystal Form: Implications for the Reaction Mechanism
Proteins: Struct.,Funct., Genet., 16, 1993
|
|
1TRD
 
 | |
2UYN
 
 | | Crystal structure of E. coli TdcF with bound 2-ketobutyrate | | Descriptor: | 2-KETOBUTYRIC ACID, PROTEIN TDCF | | Authors: | Burman, J.D, Stevenson, C.E.M, Sawers, R.G, Lawson, D.M. | | Deposit date: | 2007-04-11 | | Release date: | 2007-05-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of Escherichia Coli Tdcf, a Member of the Highly Conserved Yjgf/Yer057C/Uk114 Family.
Bmc Struct.Biol., 7, 2007
|
|
1TQ1
 
 | | Solution structure of At5g66040, a putative protein from Arabidosis Thaliana | | Descriptor: | senescence-associated family protein | | Authors: | Cornilescu, C.C, Cornilescu, G, Singh, S, Lee, M.S, Tyler, E.M, Shahan, M.N, Vinarov, D, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-06-16 | | Release date: | 2004-06-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a single-domain thiosulfate sulfurtransferase from Arabidopsis thaliana.
Protein Sci., 15, 2006
|
|
1TPD
 
 | |
2Y30
 
 | | Simocyclinone D8 bound form of TetR-like repressor SimR | | Descriptor: | CHLORIDE ION, PUTATIVE REPRESSOR SIMREG2, SIMOCYCLINONE D8 | | Authors: | Le, T.B.K, Stevenson, C.E.M, Fiedler, H.-P, Maxwell, A, Lawson, D.M, Buttner, M.J. | | Deposit date: | 2010-12-17 | | Release date: | 2011-03-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of the Tetr-Like Simocyclinone Efflux Pump Repressor, Simr, and the Mechanism of Ligand-Mediated Derepression.
J.Mol.Biol., 408, 2011
|
|
2Y3P
 
 | | Crystal structure of N-terminal domain of GyrA with the antibiotic simocyclinone D8 | | Descriptor: | DNA GYRASE SUBUNIT A, MAGNESIUM ION, SIMOCYCLINONE D8 | | Authors: | Edwards, M.J, Flatman, R.H, Mitchenall, L.A, Stevenson, C.E.M, Le, T.B.K, Clarke, T.A, McKay, A.R, Fiedler, H.-P, Buttner, M.J, Lawson, D.M, Maxwell, A. | | Deposit date: | 2010-12-22 | | Release date: | 2010-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | A Crystal Structure of the Bifunctional Antibiotic Simocyclinone D8, Bound to DNA Gyrase.
Science, 326, 2009
|
|
1UW8
 
 | | CRYSTAL STRUCTURE OF OXALATE DECARBOXYLASE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MANGANESE (II) ION, OXALATE DECARBOXYLASE OXDC | | Authors: | Just, V.J, Stevenson, C.E.M, Bowater, L, Tanner, A, Lawson, D.M, Bornemann, S. | | Deposit date: | 2004-02-02 | | Release date: | 2004-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Closed Conformation of Bacillus Subtilis Oxalate Decarboxylase Oxdc Provides Evidence for the True Identity of the Active Site
J.Biol.Chem., 279, 2004
|
|
1SIZ
 
 | |
1SY1
 
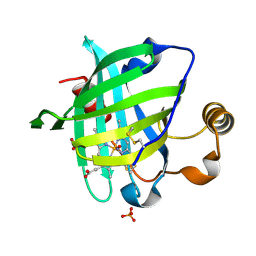 | | 1.0 A Crystal Structure of T121V Mutant of Nitrophorin 4 Complexed with Nitric Oxide | | Descriptor: | NITRIC OXIDE, Nitrophorin 4, PHOSPHATE ION, ... | | Authors: | Maes, E.M, Weichsel, A, Andersen, J.F, Shepley, D, Montfort, W.R. | | Deposit date: | 2004-03-31 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Role of binding site loops in controlling nitric oxide release: structure and kinetics of mutant forms of nitrophorin 4
Biochemistry, 43, 2004
|
|
1W6C
 
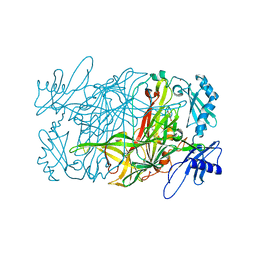 | | AGAO holoenzyme in a small cell, at 2.2 angstroms | | Descriptor: | COPPER (II) ION, PHENYLETHYLAMINE OXIDASE, SODIUM ION | | Authors: | Duff, A.P, Langley, D.B, Juda, G.A, Shepard, E.M, Dooley, D.M, Freeman, H.C, Guss, J.M. | | Deposit date: | 2004-08-17 | | Release date: | 2005-12-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Copper Containing Amine Oxidase from Arthrobacter Globiformis: Refinement at 1.55 And 2.20 A Resolution in Two Crystal Forms.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
1VJM
 
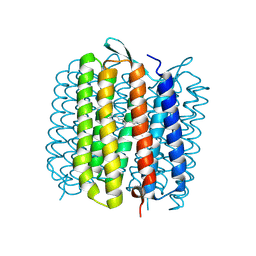 | | Deformation of helix C in the low-temperature L-intermediate of bacteriorhodopsin | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Edman, K, Royant, A, Larsson, G, Jacobson, F, Taylor, T, van der Spoel, D, Landau, E.M, Pebay-Peyroula, E, Neutze, R. | | Deposit date: | 2004-03-12 | | Release date: | 2004-04-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Deformation of helix C in the low temperature L-intermediate of bacteriorhodopsin.
J.Biol.Chem., 279, 2004
|
|
1VKG
 
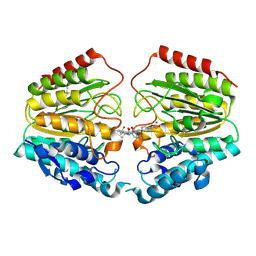 | | Crystal Structure of Human HDAC8 complexed with CRA-19156 | | Descriptor: | 5-(4-METHYL-BENZOYLAMINO)-BIPHENYL-3,4'-DICARBOXYLIC ACID 3-DIMETHYLAMIDE-4'-HYDROXYAMIDE, Histone deacetylase 8, SODIUM ION, ... | | Authors: | Somoza, J.R, Skene, R.J, Katz, B.A, Mol, C, Ho, J.D, Jennings, A.J, Luong, C, Arvai, A, Buggy, J.J, Chi, E, Tang, J, Sang, B.-C, Verner, E, Wynands, R, Leahy, E.M, Dougan, D.R, Snell, G, Navre, M, Knuth, M.W, Swanson, R.V, McRee, D.E, Tari, L.W. | | Deposit date: | 2004-05-13 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Snapshots of Human HDAC8 Provide Insights into the Class I Histone Deacetylases
Structure, 12, 2004
|
|
1W2Z
 
 | | PSAO and Xenon | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AMINE OXIDASE, COPPER CONTAINING, ... | | Authors: | Duff, A.P, Trambaiolo, D.M, Cohen, A.E, Ellis, P.J, Juda, G.A, Shepard, E.M, Langley, D.B, Dooley, D.M, Freeman, H.C, Guss, J.M. | | Deposit date: | 2004-07-11 | | Release date: | 2004-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Using Xenon as a Probe for Dioxygen-Binding Sites in Copper Amine Oxidases.
J.Mol.Biol., 344, 2004
|
|
4PWA
 
 | |
4WMG
 
 | | Structure of hen egg-white lysozyme from a microfludic harvesting device using synchrotron radiation (2.5A) | | Descriptor: | Lysozyme C | | Authors: | Lyubimov, A.Y, Murray, T.D, Koehl, A, Uervirojnangkoorn, M, Zeldin, O.B, Cohen, A.E, Soltis, S.M, Baxter, E.M, Brewster, A.S, Sauter, N.K, Brunger, A.T, Berger, J.M. | | Deposit date: | 2014-10-08 | | Release date: | 2015-04-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Capture and X-ray diffraction studies of protein microcrystals in a microfluidic trap array.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4ZFI
 
 | | Structure of Mdm2 with low molecular weight inhibitor | | Descriptor: | (5S)-3,5-bis(4-chlorobenzyl)-4-(6-chloro-1H-indol-3-yl)-5-hydroxy-1-methyl-1,5-dihydro-2H-pyrrol-2-one, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Zak, K.M, Twarda-Clapa, A, Wrona, E.M, Grudnik, P, Dubin, G, Holak, T.A. | | Deposit date: | 2015-04-21 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Unique Mdm2-Binding Mode of the 3-Pyrrolin-2-one- and 2-Furanone-Based Antagonists of the p53-Mdm2 Interaction.
ACS Chem. Biol., 11, 2016
|
|
4ZGK
 
 | | Structure of Mdm2 with low molecular weight inhibitor. | | Descriptor: | (5R)-3,5-bis(4-chlorobenzyl)-4-(6-chloro-1H-indol-3-yl)-5-hydroxyfuran-2(5H)-one, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Twarda-Clapa, A, Zak, K.M, Wrona, E.M, Grudnik, P, Dubin, G, Holak, T.A. | | Deposit date: | 2015-04-23 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Unique Mdm2-Binding Mode of the 3-Pyrrolin-2-one- and 2-Furanone-Based Antagonists of the p53-Mdm2 Interaction.
ACS Chem. Biol., 11, 2016
|
|
8A3N
 
 | | Geissoschizine synthase from Catharanthus roseus - binary complex with NADP+ | | Descriptor: | Geissoschizine synthase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ZINC ION | | Authors: | Langley, C, Tatsis, E, Hong, B, Nakamura, Y, Kamileen, M.O, Paetz, C, Stevenson, C.E.M, Basquin, J, Lawson, D.M, Caputi, L, O'Connor, S.E. | | Deposit date: | 2022-06-08 | | Release date: | 2022-10-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Expansion of the Catalytic Repertoire of Alcohol Dehydrogenases in Plant Metabolism.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
8AV8
 
 | | Small molecular stabilizer for ERalpha and 14-3-3 (1075300) | | Descriptor: | 14-3-3 protein sigma, 2-chloranyl-~{N}-[[1-(1-phenoxycyclopentyl)carbonylpiperidin-4-yl]methyl]ethanamide, Estrogen receptor, ... | | Authors: | Visser, E.J, Vandenboorn, E.M.F, Ottmann, C. | | Deposit date: | 2022-08-26 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14-3-3 sigma /ER alpha Protein-Protein Interaction from Nonselective Fragments.
J.Am.Chem.Soc., 145, 2023
|
|
8ANF
 
 | | Small molecule stabilizer for ERalpha and 14-3-3 (1074359) | | Descriptor: | 14-3-3 protein sigma, 2-chloranyl-N-[3-[1-[2-(4-chloranylphenoxy)-2-methyl-propanoyl]piperidin-4-yl]propyl]ethanamide, Estrogen receptor, ... | | Authors: | Konstantinidou, M, Visser, E.J, Vandenboorn, E.M.F, Sheng, C, Jaishankar, P, Overmans, M.J.A.M, Dutta, S, Neitz, J, Renslo, A, Ottmann, C, Brunsveld, L, Arkin, M. | | Deposit date: | 2022-08-05 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14-3-3 sigma /ER alpha Protein-Protein Interaction from Nonselective Fragments.
J.Am.Chem.Soc., 145, 2023
|
|
8AQC
 
 | | Small molecular stabilizer for ERalpha and 14-3-3 (1080294) | | Descriptor: | 14-3-3 protein sigma, Estrogen receptor, MAGNESIUM ION, ... | | Authors: | Visser, E.J, Vandenboorn, E.M.F, Ottmann, C. | | Deposit date: | 2022-08-12 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14-3-3 sigma /ER alpha Protein-Protein Interaction from Nonselective Fragments.
J.Am.Chem.Soc., 145, 2023
|
|
8ARW
 
 | | Small molecular stabilizer for ERalpha and 14-3-3 (1076402) | | Descriptor: | 14-3-3 protein sigma, Estrogen receptor, MAGNESIUM ION, ... | | Authors: | Visser, E.J, Vandenboorn, E.M.F, Ottmann, C. | | Deposit date: | 2022-08-17 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14-3-3 sigma /ER alpha Protein-Protein Interaction from Nonselective Fragments.
J.Am.Chem.Soc., 145, 2023
|
|
8ARY
 
 | | Small molecular stabilizer for ERalpha and 14-3-3 (1080273) | | Descriptor: | 14-3-3 protein sigma, 2-chloranyl-N-[[1-[1-(4-chloranylphenoxy)cyclohexyl]carbonylpiperidin-4-yl]methyl]ethanamide, Estrogen receptor, ... | | Authors: | Visser, E.J, Vandenboorn, E.M.F, Ottmann, C. | | Deposit date: | 2022-08-17 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14-3-3 sigma /ER alpha Protein-Protein Interaction from Nonselective Fragments.
J.Am.Chem.Soc., 145, 2023
|
|
