8Q8K
 
 | | KI Polyomavirus LTA NLS bound to importin alpha 2 | | Descriptor: | Importin subunit alpha-1, Large T antigen | | Authors: | Cross, E.M, Forwood, J.K, Alvisi, G. | | Deposit date: | 2023-08-18 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A functional and structural comparative analysis of large tumor antigens reveals evolution of different importin alpha-dependent nuclear localization signals.
Protein Sci., 33, 2024
|
|
7NGB
 
 | | Structure of Wild-Type Human Potassium Chloride Transporter KCC3 in NaCl (LMNG/CHS) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform 2 of Solute carrier family 12 member 6, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Chi, G, Man, H, Pike, A.C.W, Wang, D, McKinley, G, Mukhopadhyay, S.M.M, MacLean, E.M, Chalk, R, Moreau, C, Snee, M, Abrusci, P, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Marsden, B.D, Burgess-Brown, N.A, Duerr, K.L. | | Deposit date: | 2021-02-09 | | Release date: | 2021-06-09 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Phospho-regulation, nucleotide binding and ion access control in potassium-chloride cotransporters.
Embo J., 40, 2021
|
|
7N8J
 
 | |
8DDI
 
 | |
8DDM
 
 | |
8SDW
 
 | | Crystal structure of the non-myristoylated mutant [L8K]Arf1 in complex with a GDP analogue | | Descriptor: | ADP-ribosylation factor 1, GUANOSINE-3'-MONOPHOSPHATE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Rosenberg Jr, E.M, Randazzo, P.A, Esser, L, Xia, D. | | Deposit date: | 2023-04-07 | | Release date: | 2023-06-28 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Point mutations in Arf1 reveal cooperative effects of the N-terminal extension and myristate for GTPase-activating protein catalytic activity.
Plos One, 19, 2024
|
|
8DGB
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) Q192T Mutant in Complex with Inhibitor GC376 | | Descriptor: | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | Authors: | Lewandowski, E.M, Jacobs, L.M.C, Hu, Y, Tan, H, Wang, J, Chen, Y. | | Deposit date: | 2022-06-23 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
8DCZ
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) M165Y Mutant in Complex with Nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Lewandowski, E.M, Hu, Y, Tan, H, Wang, J, Chen, Y. | | Deposit date: | 2022-06-17 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
8D4L
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) S144A Mutant | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Lewandowski, E.M, Hu, Y, Tan, H, Wang, J, Chen, Y. | | Deposit date: | 2022-06-02 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir.
Biorxiv, 2022
|
|
8DD1
 
 | | SARS-CoV-2 Main Protease (Mpro) H164N Mutant in Complex with Inhibitor GC376 | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | Authors: | Lewandowski, E.M, Butler, S.G, Hu, Y, Tan, H, Wang, J, Chen, Y. | | Deposit date: | 2022-06-17 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
6UUT
 
 | |
8QXX
 
 | | HCMV DNA polymerase processivity factor UL44 phosphorylated NLS 410-433 bound to mouse importin alpha 2 | | Descriptor: | DNA polymerase processivity factor, Importin subunit alpha-1 | | Authors: | Cross, E.M, Marin, O, Ariawan, D, Aragao, D, Cozza, G, Di Iorio, E, Forwood, J.K, Alvisi, G. | | Deposit date: | 2023-10-25 | | Release date: | 2023-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural determinants of phosphorylation-dependent nuclear transport of HCMV DNA polymerase processivity factor UL44.
Febs Lett., 598, 2024
|
|
8QXW
 
 | | HCMV DNA polymerase processivity factor UL44 unphosphorylated NLS 410-433 bound to mouse importin alpha 2 | | Descriptor: | DNA polymerase processivity factor, Importin subunit alpha-1 | | Authors: | Cross, E.M, Marin, O, Ariawan, D, Aragao, D, Cozza, G, Di Iorio, E, Forwood, J.K, Alvisi, G. | | Deposit date: | 2023-10-25 | | Release date: | 2023-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural determinants of phosphorylation-dependent nuclear transport of HCMV DNA polymerase processivity factor UL44.
Febs Lett., 598, 2024
|
|
3T1Y
 
 | | Structure of the Thermus thermophilus 30S ribosomal subunit complexed with a human anti-codon stem loop (HASL) of transfer RNA Lysine 3 (TRNALYS3) bound to an mRNA with an AAG-codon in the A-site and paromomycin | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Murphy, F.V, Vendeix, F.A.P, Cantara, W, Leszczynska, G, Gustilo, E.M, Sproat, B, Malkiewicz, A.A.P, Agris, P.F. | | Deposit date: | 2011-07-22 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Human tRNA(Lys3)(UUU) Is Pre-Structured by Natural Modifications for Cognate and Wobble Codon Binding through Keto-Enol Tautomerism.
J.Mol.Biol., 416, 2012
|
|
6V3T
 
 | |
8ROZ
 
 | |
6VAU
 
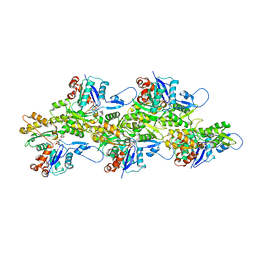 | | Bare actin filament from a partially cofilin-decorated sample | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Huehn, A.R, Bibeau, J.P, Schramm, A.C, Cao, W, De La Cruz, E.M, Sindelar, C.V. | | Deposit date: | 2019-12-17 | | Release date: | 2020-01-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of cofilin-induced structural changes reveal local and asymmetric perturbations of actin filaments.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3T1H
 
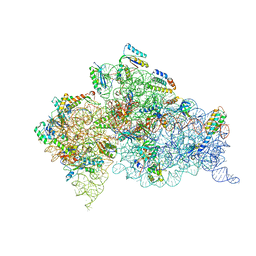 | | Structure of the Thermus thermophilus 30S ribosomal subunit complexed with a human anti-codon stem loop (HASL) of transfer RNA lysine 3 (tRNALys3) bound to an mRNA with an AAA-codon in the A-site and Paromomycin | | Descriptor: | 16s rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Murphy, F.V, Vendeix, F.A.P, Cantara, W, Leszczynska, G, Gustilo, E.M, Sproat, B, Malkiewicz, A.A.P, Agris, P.F. | | Deposit date: | 2011-07-21 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Human tRNA(Lys3)(UUU) Is Pre-Structured by Natural Modifications for Cognate and Wobble Codon Binding through Keto-Enol Tautomerism.
J.Mol.Biol., 416, 2012
|
|
6VAO
 
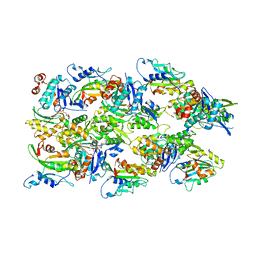 | | Human cofilin-1 decorated actin filament | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Huehn, A.R, Bibeau, J.P, Schramm, A.C, Cao, W, De La Cruz, E.M, Sindelar, C.V. | | Deposit date: | 2019-12-17 | | Release date: | 2020-01-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of cofilin-induced structural changes reveal local and asymmetric perturbations of actin filaments.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VGJ
 
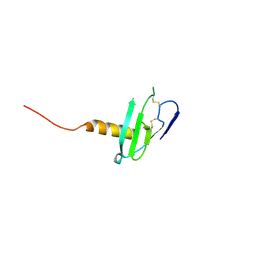 | | N-terminal variant of CXCL13 | | Descriptor: | C-X-C motif chemokine 13 | | Authors: | Rosenberg Jr, E.M, Lolis, E.J. | | Deposit date: | 2020-01-08 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | The N-terminal length and side-chain composition of CXCL13 affect crystallization, structure and functional activity.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
8SPJ
 
 | |
8SXO
 
 | |
6VPP
 
 | | Cryo-EM structure of microtubule-bound KLP61F motor with tail domain in the nucleotide-free state | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-like protein Klp61F, ... | | Authors: | Bodrug, T, Wilson-Kubalek, E.M, Nithianantham, S, Debs, G, Sindelar, C.V, Milligan, R, Al-Bassam, J. | | Deposit date: | 2020-02-04 | | Release date: | 2020-02-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | The kinesin-5 tail domain directly modulates the mechanochemical cycle of the motor domain for anti-parallel microtubule sliding.
Elife, 9, 2020
|
|
6VNU
 
 | | X-ray Crystal Structure of Ruthenocenyl-7-Aminocephalosporanic Acid Covalent Acyl-Enzyme Complex with CTX-M-14 E166A Beta-Lactamase | | Descriptor: | Beta-lactamase, POTASSIUM ION, [(1,2,3,4,5-eta)-1-(4-{[carboxy(4-carboxy-5-methylidene-5,6-dihydro-2H-1,3-thiazin-2-yl)methyl]amino}-4-oxobutanoyl)cyclopentadienyl][(1,2,3,4,5-eta)-cyclopentadienyl]ruthenium, ... | | Authors: | Lewandowski, E.M, Jacobs, L.M.C, Chen, Y. | | Deposit date: | 2020-01-29 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Metallocenyl 7-ACA Conjugates: Antibacterial Activity Studies and Atomic-Resolution X-ray Crystal Structure with CTX-M beta-Lactamase.
Chembiochem, 21, 2020
|
|
3R92
 
 | | Discovery of a macrocyclic o-aminobenzamide Hsp90 inhibitor with heterocyclic tether that shows extended biomarker activity and in vivo efficacy in a mouse xenograft model. | | Descriptor: | (3aR)-13,13,16-trimethyl-15-oxo-1,2,3,3a,4,5,12,14,15,17,18,19-dodecahydro-13H-10,6-(metheno)pyrrolo[2',1':3,4][1,4,9]triazacyclotetradecino[9,8-a]indole-7-carboxamide, Heat shock protein HSP 90-alpha | | Authors: | Zapf, C.W, Bloom, J.D, McBean, J.L, Dushin, R.G, Golas, J.M, Liu, H, Lucas, J, Boschelli, F, Vogan, E.M, Levin, J.I. | | Deposit date: | 2011-03-24 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5801 Å) | | Cite: | Discovery of a macrocyclic o-aminobenzamide Hsp90 inhibitor with heterocyclic tether that shows extended biomarker activity and in vivo efficacy in a mouse xenograft model.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
