8SG5
 
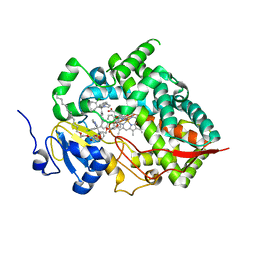 | | Cytochrome P450 (CYP) 3A5 crystallized with clotrimazole | | Descriptor: | 1-[(2-CHLOROPHENYL)(DIPHENYL)METHYL]-1H-IMIDAZOLE, Cytochrome P450 3A5, DODECYL NONA ETHYLENE GLYCOL ETHER, ... | | Authors: | Hsu, M.H, Johnson, E.F. | | Deposit date: | 2023-04-11 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Differential Effects of Clotrimazole on X-Ray Crystal Structures of Human Cytochromes P450 3A5 and 3A4.
Drug Metab.Dispos., 51, 2023
|
|
8SPD
 
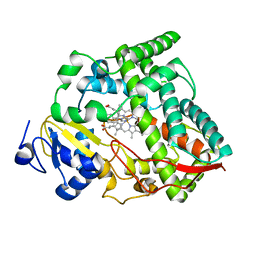 | | Cytochrome P450 (CYP) 3A4 crystallized with clotrimazole | | Descriptor: | 1-[(2-CHLOROPHENYL)(DIPHENYL)METHYL]-1H-IMIDAZOLE, Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hsu, M.H, Johnson, E.F. | | Deposit date: | 2023-05-02 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Differential Effects of Clotrimazole on X-Ray Crystal Structures of Human Cytochromes P450 3A5 and 3A4.
Drug Metab.Dispos., 51, 2023
|
|
7LVE
 
 | |
7LVG
 
 | |
7LVF
 
 | |
1N6B
 
 | | Microsomal Cytochrome P450 2C5/3LVdH Complex with a dimethyl derivative of sulfaphenazole | | Descriptor: | 4-METHYL-N-METHYL-N-(2-PHENYL-2H-PYRAZOL-3-YL)BENZENESULFONAMIDE, Cytochrome P450 2C5, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Wester, M.R, Johnson, E.F, Marques-Soares, C, Dansette, P.M, Mansuy, D, Stout, C.D. | | Deposit date: | 2002-11-09 | | Release date: | 2003-06-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a Substrate Complex of Mammalian Cytochrome P450 2C5 at 2.3 A Resolution: Evidence
for Multiple Substrate Binding Modes
Biochemistry, 42, 2003
|
|
4PNS
 
 | | Crystal Structure of human Tankyrase 2 in complex with INH2BP. | | Descriptor: | 6-amino-5-iodo-2H-chromen-2-one, GLYCEROL, Tankyrase-2, ... | | Authors: | Qiu, W, Lam, R, Romanov, V, Gordon, R, Gebremeskel, S, Vodsedalek, J, Thompson, C, Beletskaya, I, Battaile, K.P, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-05-25 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Insights into the binding of PARP inhibitors to the catalytic domain of human tankyrase-2.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4PNN
 
 | | Crystal Structure of human Tankyrase 2 in complex with 4HQN. | | Descriptor: | Tankyrase-2, ZINC ION, quinazolin-4(1H)-one | | Authors: | Qiu, W, Lam, R, Romanov, V, Gordon, R, Gebremeskel, S, Vodsedalek, J, Thompson, C, Beletskaya, I, Battaile, K.P, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-05-24 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Insights into the binding of PARP inhibitors to the catalytic domain of human tankyrase-2.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4PEO
 
 | | Crystal structure of a hypothetical protein from Staphylococcus aureus. | | Descriptor: | Hypothetical protein | | Authors: | McGrath, T.E, Kisselman, G, Romanov, V, Wu-Brown, J, Soloveychik, M, Chan, T.S.Y, Gordon, R.D, Thambipillai, D, Dharamsi, A, Mansoury, K, Battaile, K.P, Edwards, A.M, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-04-24 | | Release date: | 2015-05-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of a hypothetical protein from Staphylococcus aureus.
To Be Published
|
|
4PML
 
 | | Crystal Structure of human Tankyrase 2 in complex with 3-amino-benzamide. | | Descriptor: | 1,2-ETHANEDIOL, 3-aminobenzamide, DIMETHYL SULFOXIDE, ... | | Authors: | Qiu, W, Lam, R, Romanov, V, Gordon, R, Gebremeskel, S, Vodsedalek, J, Thompson, C, Beletskaya, I, Battaile, K.P, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-05-22 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Insights into the binding of PARP inhibitors to the catalytic domain of human tankyrase-2.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4PNL
 
 | | Crystal structure of TNKS-2 in complex with DR2313. | | Descriptor: | 1,2-ETHANEDIOL, 2-methyl-3,5,7,8-tetrahydro-4H-thiopyrano[4,3-d]pyrimidin-4-one, DIMETHYL SULFOXIDE, ... | | Authors: | Qiu, W, Lam, R, Romanov, V, Gordon, R, Gebremeskel, S, Vodsedalek, J, Thompson, C, Beletskaya, I, Battaile, K.P, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-05-23 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Insights into the binding of PARP inhibitors to the catalytic domain of human tankyrase-2.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4PAU
 
 | | Hypothetical protein SA1058 from S. aureus. | | Descriptor: | CHLORIDE ION, Nitrogen regulatory protein A | | Authors: | Battaile, K.P, Wu-Brown, J, Romanov, V, Jones, K, Lam, R, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-04-10 | | Release date: | 2015-05-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Hypothetical protein SA1058 from S. aureus.
To Be Published
|
|
4PNM
 
 | | Crystal Structure of human Tankyrase 2 in complex with Nu1025. | | Descriptor: | 1,2-ETHANEDIOL, 8-HYDROXY-2-METHYL-3-HYDRO-QUINAZOLIN-4-ONE, Tankyrase-2, ... | | Authors: | Qiu, W, Lam, R, Romanov, V, Gordon, R, Gebremeskel, S, Vodsedalek, J, Thompson, C, Beletskaya, I, Battaile, K.P, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-05-23 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Insights into the binding of PARP inhibitors to the catalytic domain of human tankyrase-2.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4PTT
 
 | | Crystal Structure of anti-23F strep Fab C05 | | Descriptor: | ACETATE ION, Antibody pn132p2C05, heavy chain, ... | | Authors: | Bryson, S, Risnes, L, Damgupta, S, Thomson, C.A, Smith, K, Schrader, J.W, Pai, E.F. | | Deposit date: | 2014-03-11 | | Release date: | 2015-03-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of Preferred Human IgV Genes-Based Protective Antibodies Identify How Conserved Residues Contact Diverse Antigens and Assign Source of Specificity to CDR3 Loop Variation.
J. Immunol., 196, 2016
|
|
4PNQ
 
 | | Crystal Structure of human Tankyrase 2 in complex with 5AIQ. | | Descriptor: | 5-aminoisoquinolin-1(4H)-one, ISOPROPYL ALCOHOL, Tankyrase-2, ... | | Authors: | Qiu, W, Lam, R, Romanov, V, Gordon, R, Gebremeskel, S, Vodsedalek, J, Thompson, C, Beletskaya, I, Battaile, K.P, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-05-24 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Insights into the binding of PARP inhibitors to the catalytic domain of human tankyrase-2.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7SV2
 
 | | Human Cytochrome P450 (CYP) 3A5 ternary complex with azamulin | | Descriptor: | (3aS,4R,5S,6R,8R,9R,9aR,10R)-6-ethyl-5-hydroxy-4,6,9,10-tetramethyl-1-oxodecahydro-3a,9-propanocyclopenta[8]annulen-8-yl [(5-amino-1H-1,2,4-triazol-3-yl)sulfanyl]acetate, Cytochrome P450 3A5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hsu, M, Johnson, E.F. | | Deposit date: | 2021-11-18 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structural characterization of the homotropic cooperative binding of azamulin to human cytochrome P450 3A5.
J.Biol.Chem., 298, 2022
|
|
4PNR
 
 | | Crystal Structure of human Tankyrase 2 in complex with TIQ-A. | | Descriptor: | 4H-thieno[2,3-c]isoquinolin-5-one, ETHANOL, GLYCEROL, ... | | Authors: | Qiu, W, Lam, R, Romanov, V, Gordon, R, Gebremeskel, S, Vodsedalek, J, Thompson, C, Beletskaya, I, Battaile, K.P, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-05-24 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Insights into the binding of PARP inhibitors to the catalytic domain of human tankyrase-2.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4PTU
 
 | | Crystal Structure of anti-23F strep Fab C05 with rhamnose | | Descriptor: | ACETATE ION, Antibody pn132p2C05, heavy chain, ... | | Authors: | Bryson, S, Risnes, L, Damgupta, S, Thomson, C.A, Smith, K, Schrader, J.W, Pai, E.F. | | Deposit date: | 2014-03-11 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.511 Å) | | Cite: | Structures of Preferred Human IgV Genes-Based Protective Antibodies Identify How Conserved Residues Contact Diverse Antigens and Assign Source of Specificity to CDR3 Loop Variation.
J. Immunol., 196, 2016
|
|
4PNT
 
 | | Crystal Structure of human Tankyrase 2 in complex with 1,5-IQD. | | Descriptor: | 5-hydroxyisoquinolin-1(4H)-one, Tankyrase-2, ZINC ION | | Authors: | Qiu, W, Lam, R, Romanov, V, Gordon, R, Gebremeskel, S, Vodsedalek, J, Thompson, C, Beletskaya, I, Battaile, K.P, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-05-25 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insights into the binding of PARP inhibitors to the catalytic domain of human tankyrase-2.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5K3A
 
 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - His280Asn/Fluoroacetate - Cocrystallized - Both Protomers Reacted with Ligand | | Descriptor: | CHLORIDE ION, Fluoroacetate dehalogenase | | Authors: | Mehrabi, P, Kim, T.H, Prosser, S.R, Pai, E.F. | | Deposit date: | 2016-05-19 | | Release date: | 2017-02-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.511 Å) | | Cite: | The role of dimer asymmetry and protomer dynamics in enzyme catalysis.
Science, 355, 2017
|
|
5K3B
 
 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - Asp110Asn/Chloroacetate - Cocrystallized | | Descriptor: | CHLORIDE ION, Fluoroacetate dehalogenase, chloroacetic acid | | Authors: | Mehrabi, P, Kim, T.H, Prosser, S.R, Pai, E.F. | | Deposit date: | 2016-05-19 | | Release date: | 2017-02-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The role of dimer asymmetry and protomer dynamics in enzyme catalysis.
Science, 355, 2017
|
|
5K3D
 
 | |
5K3C
 
 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - WT/5-Fluorotryptophan | | Descriptor: | CHLORIDE ION, Fluoroacetate dehalogenase | | Authors: | Mehrabi, P, Kim, T.H, Prosser, S.R, Pai, E.F. | | Deposit date: | 2016-05-19 | | Release date: | 2017-02-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.541 Å) | | Cite: | The role of dimer asymmetry and protomer dynamics in enzyme catalysis.
Science, 355, 2017
|
|
5K3F
 
 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - His280Asn/Fluoroacetate - Cocrystallized - Single Protomer Reacted with Ligand | | Descriptor: | CHLORIDE ION, Fluoroacetate dehalogenase | | Authors: | Mehrabi, P, Kim, T.H, Prosser, S.R, Pai, E.F. | | Deposit date: | 2016-05-19 | | Release date: | 2017-02-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | The role of dimer asymmetry and protomer dynamics in enzyme catalysis.
Science, 355, 2017
|
|
5K3E
 
 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - Asp110Asn/Glycolate - Cocrystallized | | Descriptor: | CHLORIDE ION, Fluoroacetate dehalogenase, GLYCOLIC ACID | | Authors: | Mehrabi, P, Kim, T.H, Prosser, S.R, Pai, E.F. | | Deposit date: | 2016-05-19 | | Release date: | 2017-02-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | The role of dimer asymmetry and protomer dynamics in enzyme catalysis.
Science, 355, 2017
|
|
