7L5C
 
 | | Structure of copper bound MEMO1 | | Descriptor: | 1,2-ETHANEDIOL, COPPER (I) ION, GLYCEROL, ... | | Authors: | Boniecki, M.T, Uhlemann, E.E, Dmitriev, O.Y. | | Deposit date: | 2020-12-21 | | Release date: | 2022-01-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | MEMO1 binds iron and modulates iron homeostasis in cancer cells.
Elife, 13, 2024
|
|
7LKQ
 
 | | The PilZ(delta107-117)-FimX(GGDEF-EAL) complex from Xanthomonas citri | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), FimX(GGDEF-EAL), MAGNESIUM ION, ... | | Authors: | Llontop, E.E, Guzzo, C.R, Farah, C.S. | | Deposit date: | 2021-02-02 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The PilB-PilZ-FimX regulatory complex of the Type IV pilus from Xanthomonas citri.
Plos Pathog., 17, 2021
|
|
7LKO
 
 | |
7LKN
 
 | |
7LKM
 
 | |
1WS0
 
 | |
1WS1
 
 | |
5ED7
 
 | | Crystal Structure of HSV-1 UL21 C-terminal Domain | | Descriptor: | CHLORIDE ION, Tegument protein UL21 | | Authors: | Metrick, C.M, Heldwein, E.E. | | Deposit date: | 2015-10-20 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Novel Structure and Unexpected RNA-Binding Ability of the C-Terminal Domain of Herpes Simplex Virus 1 Tegument Protein UL21.
J.Virol., 90, 2016
|
|
1XV3
 
 | | NMR structure of the synthetic penaeidin 4 | | Descriptor: | Penaeidin-4d | | Authors: | Cuthbertson, B.J, Yang, Y, Bullesbach, E.E, Bachere, E, Gross, P.S, Aumelas, A. | | Deposit date: | 2004-10-27 | | Release date: | 2005-02-15 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of synthetic penaeidin-4 with structural and functional comparisons to penaeidin-3
J.Biol.Chem., 280, 2005
|
|
5GVJ
 
 | | Structure of FabK (M276A) mutant from Thermotoga maritima | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [FMN], SODIUM ION | | Authors: | Kim, E.E, Shin, S.C, Ha, B.H, Moon, J.H. | | Deposit date: | 2016-09-06 | | Release date: | 2017-04-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and biochemical characterization of FabK from Thermotoga maritima.
Biochem. Biophys. Res. Commun., 482, 2017
|
|
5GVH
 
 | | Structure of FabK from Thermotoga maritima | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [FMN], FLAVIN MONONUCLEOTIDE, SODIUM ION | | Authors: | Kim, E.E, Shin, S.C, Ha, B.H, Moon, J.H. | | Deposit date: | 2016-09-05 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.294 Å) | | Cite: | Structural and biochemical characterization of FabK from Thermotoga maritima.
Biochem. Biophys. Res. Commun., 482, 2017
|
|
5EHU
 
 | |
5H22
 
 | | Hsp90 alpha N-terminal domain in complex with an inhibitor | | Descriptor: | 4-chloranyl-7-[(4-methoxy-3,5-dimethyl-pyridin-2-yl)methyl]-5-(phenylmethyl)pyrrolo[2,3-d]pyrimidin-2-amine, Hsp90aa1 protein | | Authors: | Kim, E.E, Shin, S.C, Keum, G.C. | | Deposit date: | 2016-10-13 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Synthesis and in vitro antiproliferative activity of C5-benzyl substituted 2-amino-pyrrolo[2,3-d]pyrimidines as potent Hsp90 inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
1XVP
 
 | | crystal structure of CAR/RXR heterodimer bound with SRC1 peptide, fatty acid and CITCO | | Descriptor: | 6-(4-CHLOROPHENYL)IMIDAZO[2,1-B][1,3]THIAZOLE-5-CARBALDEHYDE O-(3,4-DICHLOROBENZYL)OXIME, Orphan nuclear receptor NR1I3, PENTADECANOIC ACID, ... | | Authors: | Xu, R.X, Lambert, M.H, Wisely, B.B, Warren, E.N, Weinert, E.E, Waitt, G.M, Williams, J.D, Moore, L.B, Willson, T.M, Moore, J.T. | | Deposit date: | 2004-10-28 | | Release date: | 2004-12-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Structural Basis for Constitutive Activity in the Human CAR/RXRalpha Heterodimer.
Mol.Cell, 16, 2004
|
|
5GKA
 
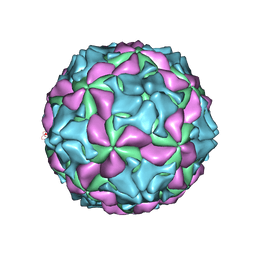 | | cryo-EM structure of human Aichi virus | | Descriptor: | Genome polyprotein, capsid protein VP0, capsid protein VP1 | | Authors: | Zhu, L, Wang, X.X, Ren, J.S, Tuthill, T.J, Fry, E.E, Rao, Z.H, Stuart, D.I. | | Deposit date: | 2016-07-04 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of human Aichi virus and implications for receptor binding
Nat Microbiol, 1, 2016
|
|
1XPN
 
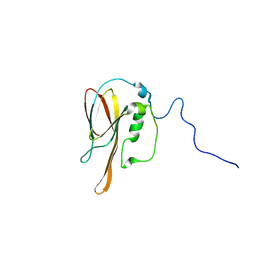 | | NMR structure of P. aeruginosa protein PA1324: Northeast Structural Genomics Consortium target PaP1 | | Descriptor: | hypothetical protein PA1324 | | Authors: | Cort, J.R, Ni, S, Lockert, E.E, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-10-08 | | Release date: | 2004-11-23 | | Last modified: | 2024-03-06 | | Method: | SOLUTION NMR | | Cite: | Structure and function of Pseudomonas aeruginosa protein PA1324 (21-170).
Protein Sci., 18, 2009
|
|
5EJJ
 
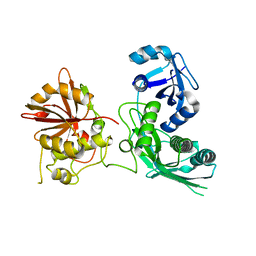 | | Crystal structure of UfSP from C.elegans | | Descriptor: | Ufm1-specific protease | | Authors: | Kim, K, Ha, B, Kim, E.E. | | Deposit date: | 2015-11-02 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The MPN domain of Caenorhabditis elegans UfSP modulates both substrate recognition and deufmylation activity
Biochem. Biophys. Res. Commun., 476, 2016
|
|
1XV9
 
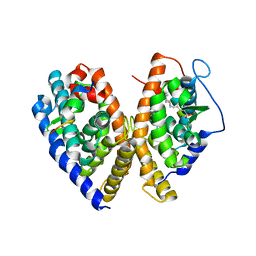 | | crystal structure of CAR/RXR heterodimer bound with SRC1 peptide, fatty acid, and 5b-pregnane-3,20-dione. | | Descriptor: | (5BETA)-PREGNANE-3,20-DIONE, Orphan nuclear receptor NR1I3, PENTADECANOIC ACID, ... | | Authors: | Xu, R.X, Lambert, M.H, Wisely, B.B, Warren, E.N, Weinert, E.E, Waitt, G.M, Williams, J.D, Moore, L.B, Willson, T.M, Moore, J.T. | | Deposit date: | 2004-10-27 | | Release date: | 2004-12-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Structural Basis for Constitutive Activity in the Human CAR/RXRalpha Heterodimer.
Mol.Cell, 16, 2004
|
|
1ZJG
 
 | | 13mer-co | | Descriptor: | 5'-D(*AP*TP*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*T)-3', 5'-D(*TP*AP*GP*CP*CP*CP*CP*GP*CP*CP*CP*CP*A)-3', COBALT HEXAMMINE(III), ... | | Authors: | Dohm, J.A, Hsu, M.H, Hwu, J.R, Huang, R.C, Moudrianakis, E.N, Lattman, E.E, Gittis, A.G. | | Deposit date: | 2005-04-28 | | Release date: | 2005-05-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Influence of Ions, Hydration, and the Transcriptional Inhibitor P4N on the Conformations of the Sp1 Binding Site.
J.Mol.Biol., 349, 2005
|
|
1ZAY
 
 | | PURINE REPRESSOR-HYPOXANTHINE-MODIFIED-PURF-OPERATOR COMPLEX | | Descriptor: | DNA (5'-D(*AP*AP*CP*GP*AP*AP*AP*AP*TP*(1AP)P*TP*TP*TP*TP*CP*GP*T)-3'), HYPOXANTHINE, PROTEIN (PURINE REPRESSOR) | | Authors: | Zheleznova, E.E, Brennan, R.G. | | Deposit date: | 1998-12-16 | | Release date: | 1999-12-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Roles of Exocyclic Groups in the Central Base-Pair Step in Modulating the Affinity of PurR for its Operator
To be Published
|
|
1ZBA
 
 | | Foot-and-Mouth Disease virus serotype A1061 complexed with oligosaccharide receptor. | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, Coat protein VP1, Coat protein VP2, ... | | Authors: | Fry, E.E, Newman, J.W, Curry, S, Najjam, S, Jackson, T, Blakemore, W, Lea, S.M, Miller, L, Burman, A, King, A.M, Stuart, D.I. | | Deposit date: | 2005-04-08 | | Release date: | 2005-06-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Foot-and-mouth disease virus serotype A1061 alone and complexed with oligosaccharide receptor: receptor conservation in the face of antigenic variation.
J.Gen.Virol., 86, 2005
|
|
1ZJF
 
 | | 12mer-spd-P4N | | Descriptor: | 5'-D(*AP*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*T)-3', 5'-D(*TP*AP*GP*CP*CP*CP*CP*GP*CP*CP*CP*C)-3' | | Authors: | Dohm, J.A, Hsu, M.H, Hwu, J.R, Huang, R.C, Moudrianakis, E.N, Lattman, E.E, Gittis, A.G. | | Deposit date: | 2005-04-28 | | Release date: | 2005-05-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Influence of Ions, Hydration, and the Transcriptional Inhibitor P4N on the Conformations of the Sp1 Binding Site.
J.Mol.Biol., 349, 2005
|
|
1ZBE
 
 | | Foot-and Mouth Disease Virus Serotype A1061 | | Descriptor: | Coat protein VP1, Coat protein VP2, Coat protein VP3, ... | | Authors: | Fry, E.E, Newman, J.W, Curry, S, Najjam, S, Jackson, T, Blakemore, W, Lea, S.M, Miller, L, Burman, A, King, A.M, Stuart, D.I. | | Deposit date: | 2005-04-08 | | Release date: | 2005-06-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of Foot-and-mouth disease virus serotype A1061 alone and complexed with oligosaccharide receptor: receptor conservation in the face of antigenic variation.
J.Gen.Virol., 86, 2005
|
|
1ZJE
 
 | | 12mer-spd | | Descriptor: | 5'-D(*AP*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*T)-3', 5'-D(*TP*AP*GP*CP*CP*CP*CP*GP*CP*CP*CP*C)-3' | | Authors: | Dohm, J.A, Hsu, M.H, Hwu, J.R, Huang, R.C, Moudrianakis, E.N, Lattman, E.E, Gittis, A.G. | | Deposit date: | 2005-04-28 | | Release date: | 2005-05-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Influence of Ions, Hydration, and the Transcriptional Inhibitor P4N on the Conformations of the Sp1 Binding Site.
J.Mol.Biol., 349, 2005
|
|
1Z0B
 
 | | Crystal Structure of A. fulgidus Lon proteolytic domain E506A mutant | | Descriptor: | CALCIUM ION, Putative protease La homolog type | | Authors: | Botos, I, Melnikov, E.E, Cherry, S, Kozlov, S, Makhovskaya, O.V, Tropea, J.E, Gustchina, A, Rotanova, T.V, Wlodawer, A. | | Deposit date: | 2005-03-01 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Atomic-resolution Crystal Structure of the Proteolytic Domain of Archaeoglobus fulgidus Lon Reveals the Conformational Variability in the Active Sites of Lon Proteases
J.Mol.Biol., 351, 2005
|
|
