3T3Z
 
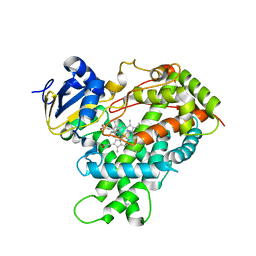 | | Human Cytochrome P450 2E1 in complex with pilocarpine | | Descriptor: | (3S,4R)-3-ethyl-4-[(1-methyl-1H-imidazol-5-yl)methyl]dihydrofuran-2(3H)-one, Cytochrome P450 2E1, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Meneely, K.M, DeVore, N.M, Scott, E.E. | | Deposit date: | 2011-07-25 | | Release date: | 2011-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural comparison of cytochromes P450 2A6, 2A13, and 2E1 with pilocarpine.
Febs J., 279, 2012
|
|
1MDN
 
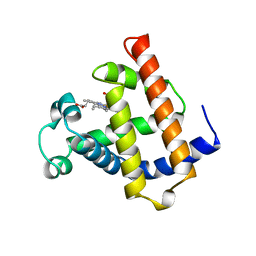 | | WILD TYPE MYOGLOBIN WITH CO | | Descriptor: | PROTEIN (MYOGLOBIN), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Murshudov, G.N, Krzywda, S, Brzozowski, A.M, Jaskolski, M, Scott, E.E, Klizas, S.A, Gibson, Q.H, Olson, J.S, Wilkinson, A.J. | | Deposit date: | 1998-08-12 | | Release date: | 1998-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Stabilizing bound O2 in myoglobin by valine68 (E11) to asparagine substitution.
Biochemistry, 37, 1998
|
|
1M6M
 
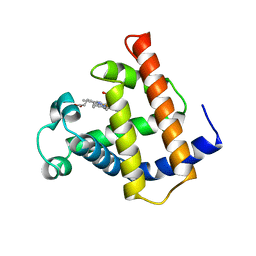 | | V68N MET MYOGLOBIN | | Descriptor: | PROTEIN (MYOGLOBIN), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Murshudov, G.N, Krzywda, S, Brzozowski, A.M, Jaskolski, M, Scott, E.E, Klizas, S.A, Gibson, Q.H, Olson, J.S, Wilkinson, A.J. | | Deposit date: | 1998-08-13 | | Release date: | 1998-08-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Stabilizing bound O2 in myoglobin by valine68 (E11) to asparagine substitution.
Biochemistry, 37, 1998
|
|
1M6C
 
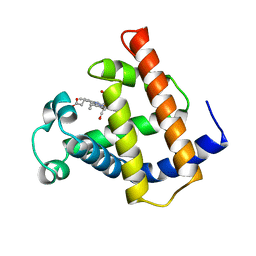 | | V68N MYOGLOBIN WITH CO | | Descriptor: | CARBON MONOXIDE, PROTEIN (MYOGLOBIN), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Murshudov, G.N, Krzywda, S, Brzozowski, A.M, Jaskolski, M, Scott, E.E, Klizas, S.A, Gibson, Q.H, Olson, J.S, Wilkinson, A.J. | | Deposit date: | 1998-08-12 | | Release date: | 1998-08-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Stabilizing bound O2 in myoglobin by valine68 (E11) to asparagine substitution.
Biochemistry, 37, 1998
|
|
1KCE
 
 | | E. COLI THYMIDYLATE SYNTHASE MUTANT E58Q IN COMPLEX WITH CB3717 AND 2'-DEOXYURIDINE 5'-MONOPHOSPHATE (DUMP) | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE | | Authors: | Sage, C.R, Rutenber, E.E, Stout, T.J, Stroud, R.M. | | Deposit date: | 1996-10-22 | | Release date: | 1997-04-21 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An essential role for water in an enzyme reaction mechanism: the crystal structure of the thymidylate synthase mutant E58Q.
Biochemistry, 35, 1996
|
|
1MWC
 
 | | WILD TYPE MYOGLOBIN WITH CO | | Descriptor: | CARBON MONOXIDE, PROTEIN (MYOGLOBIN), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Murshudov, G.N, Krzywda, S, Brzozowski, A.M, Jaskolski, M, Scott, E.E, Klizas, S.A, Gibson, Q.H, Olson, J.S, Wilkinson, A.J. | | Deposit date: | 1998-08-12 | | Release date: | 1998-08-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Stabilizing bound O2 in myoglobin by valine68 (E11) to asparagine substitution.
Biochemistry, 37, 1998
|
|
1MWD
 
 | | WILD TYPE DEOXY MYOGLOBIN | | Descriptor: | PROTEIN (MYOGLOBIN), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Murshudov, G.N, Krzywda, S, Brzozowski, A.M, Jaskolski, M, Scott, E.E, Klizas, A.A, Gibson, Q.H, Olson, J.S, Wilkinson, A.J. | | Deposit date: | 1998-08-11 | | Release date: | 1998-08-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Stabilizing bound O2 in myoglobin by valine68 (E11) to asparagine substitution.
Biochemistry, 37, 1998
|
|
1P9M
 
 | | Crystal structure of the hexameric human IL-6/IL-6 alpha receptor/gp130 complex | | Descriptor: | Interleukin-6, Interleukin-6 receptor alpha chain, Interleukin-6 receptor beta chain | | Authors: | Boulanger, M.J, Chow, D.C, Brevnova, E.E, Garcia, K.C. | | Deposit date: | 2003-05-12 | | Release date: | 2003-07-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Hexameric Structure and Assembly of the Interleukin-6/IL-6 alpha-Receptor/gp130 Complex
Science, 300, 2003
|
|
1MNO
 
 | | V68N MYOGLOBIN OXY FORM | | Descriptor: | OXYGEN MOLECULE, PROTEIN (MYOGLOBIN), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Murshudov, G.N, Krzywda, S, Brzozowski, A.M, Jaskolski, M, Scott, E.E, Klizas, S.A, Gibson, Q.H, Olson, J.S, Wilkinson, A.J. | | Deposit date: | 1998-08-13 | | Release date: | 1998-08-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Stabilizing bound O2 in myoglobin by valine68 (E11) to asparagine substitution.
Biochemistry, 37, 1998
|
|
1OOP
 
 | | The Crystal Structure of Swine Vesicular Disease Virus | | Descriptor: | Coat protein VP1, Coat protein VP2, Coat protein VP3, ... | | Authors: | Fry, E.E, Knowles, N.J, Newman, J.W.I, Wilsden, G, Rao, Z, King, A.M.Q, Stuart, D.I. | | Deposit date: | 2003-03-04 | | Release date: | 2003-04-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of Swine Vesicular Disease Virus and Implications for Host Adaptation
J.Virol., 77, 2003
|
|
1EXJ
 
 | | CRYSTAL STRUCTURE OF TRANSCRIPTION ACTIVATOR BMRR, FROM B. SUBTILIS, BOUND TO 21 BASE PAIR BMR OPERATOR AND TPP | | Descriptor: | DNA (5'-D(*AP*CP*CP*CP*TP*CP*CP*CP*CP*TP*TP*AP*GP*GP*GP*GP*AP*GP*GP*GP*T)-3'), MULTIDRUG-EFFLUX TRANSPORTER REGULATOR, SODIUM ION, ... | | Authors: | Zheleznova-Heldwein, E.E, Brennan, R.G. | | Deposit date: | 2000-05-02 | | Release date: | 2001-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the transcription activator BmrR bound to DNA and a drug.
Nature, 409, 2001
|
|
1EXI
 
 | | CRYSTAL STRUCTURE OF TRANSCRIPTION ACTIVATOR BMRR, FROM B. SUBTILIS, BOUND TO 21 BASE PAIR BMR OPERATOR AND TPSB | | Descriptor: | DNA (5'-D(*AP*CP*CP*CP*TP*CP*CP*CP*CP*TP*TP*AP*GP*GP*GP*GP*AP*GP*GP*GP*T)-3'), MULTIDRUG-EFFLUX TRANSPORTER REGULATOR, TETRAPHENYLANTIMONIUM ION, ... | | Authors: | Zheleznova-Heldwein, E.E, Brennan, R.G. | | Deposit date: | 2000-05-02 | | Release date: | 2001-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Crystal structure of the transcription activator BmrR bound to DNA and a drug.
Nature, 409, 2001
|
|
1BOW
 
 | | MULTIDRUG-BINDING DOMAIN OF TRANSCRIPTION ACTIVATOR BMRR (APO FORM) | | Descriptor: | MANGANESE (II) ION, MULTIDRUG-EFFLUX TRANSPORTER 1 REGULATOR BMRR | | Authors: | Zheleznova, E.E, Markham, P.N, Neyfakh, A.A, Brennan, R.G. | | Deposit date: | 1998-08-06 | | Release date: | 1999-08-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of multidrug recognition by BmrR, a transcription activator of a multidrug transporter.
Cell(Cambridge,Mass.), 96, 1999
|
|
7Z3Z
 
 | | Locked Wuhan SARS-CoV2 Prefusion Spike ectodomain with lipid bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, STEARIC ACID, ... | | Authors: | Duyvesteyn, H.M.E, Carrique, L, Ren, J, Stuart, D.I, Fry, E.E. | | Deposit date: | 2022-03-03 | | Release date: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The SARS-CoV-2 Spike harbours a lipid binding pocket which modulates stability of the prefusion trimer
bioRxiv, 2020
|
|
2KU0
 
 | | Inhibitor Induced Structural Change in the HCV IRES Domain IIa RNA | | Descriptor: | (7S)-7-[(dimethylamino)methyl]-1-[3-(dimethylamino)propyl]-7,8-dihydro-1H-furo[3,2-e]benzimidazol-2-amine, HCV IRES Domain IIa RNA | | Authors: | Paulsen, R.B, Seth, P.P, Swayze, E.E, Griffey, R.H, Skalicky, J.J, Cheatham III, T.E, Davis, D.R. | | Deposit date: | 2010-02-10 | | Release date: | 2010-04-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Inhibitor-induced structural change in the HCV IRES domain IIa RNA.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2KNE
 
 | | Calmodulin wraps around its binding domain in the plasma membrane CA2+ pump anchored by a novel 18-1 motif | | Descriptor: | ATPase, Ca++ transporting, plasma membrane 4, ... | | Authors: | Juranic, N, Atanasova, E, Filoteo, A.G, Macura, S, Prendergast, F.G, Penniston, J.T, Strehler, E.E. | | Deposit date: | 2009-08-21 | | Release date: | 2009-11-24 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Calmodulin wraps around its binding domain in the plasma membrane Ca2+ pump anchored by a novel 18-1 motif.
J.Biol.Chem., 285, 2010
|
|
1R9K
 
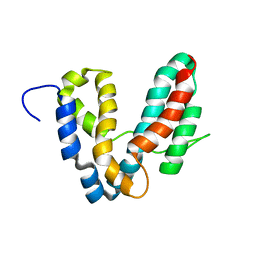 | | Representative solution structure of the catalytic domain of SopE2 | | Descriptor: | TypeIII-secreted protein effector: invasion-associated protein | | Authors: | Williams, C, Galyov, E.E, Bagby, S. | | Deposit date: | 2003-10-30 | | Release date: | 2004-09-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure, Backbone Dynamics, and Interaction with Cdc42 of Salmonella Guanine Nucleotide Exchange Factor SopE2(,).
Biochemistry, 43, 2004
|
|
2MIU
 
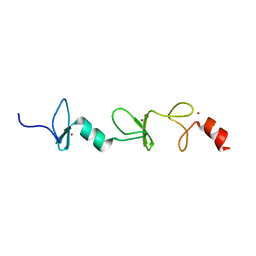 | | Structure of FHL2 LIM adaptor and its Interaction with Ski | | Descriptor: | Four and a half LIM domains protein 2, ZINC ION | | Authors: | Yang, Y, Sun, Y, Medrano, E.E, Tian, X, Weiss, M.A. | | Deposit date: | 2013-12-20 | | Release date: | 2014-01-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of FHL2 LIM adaptor and its Interaction with Ski
To be Published
|
|
2JOL
 
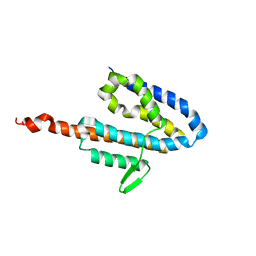 | | Average NMR structure of the catalytic domain of guanine nucleotide exchange factor BopE from Burkholderia pseudomallei | | Descriptor: | Putative G-nucleotide exchange factor | | Authors: | Wu, H, Upadhyay, A, Williams, C, Galyov, E.E, van den Elsen, J.M.H, Bagby, S. | | Deposit date: | 2007-03-14 | | Release date: | 2007-03-27 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | The guanine-nucleotide-exchange factor BopE from Burkholderia pseudomallei adopts a compact version of the Salmonella SopE/SopE2 fold and undergoes a closed-to-open conformational change upon interaction with Cdc42
Biochem.J., 411, 2008
|
|
2JOK
 
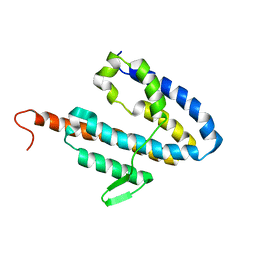 | | NMR structure of the catalytic domain of guanine nucleotide exchange factor BopE from Burkholderia pseudomallei | | Descriptor: | Putative G-nucleotide exchange factor | | Authors: | Wu, H, Upadhyay, A, Williams, C, Galyov, E.E, van den Elsen, J.M.H, Bagby, S. | | Deposit date: | 2007-03-14 | | Release date: | 2007-09-18 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | The guanine-nucleotide-exchange factor BopE from Burkholderia pseudomallei adopts a compact version of the Salmonella SopE/SopE2 fold and undergoes a closed-to-open conformational change upon interaction with Cdc42
Biochem.J., 411, 2008
|
|
1SNM
 
 | |
1PO5
 
 | | Structure of mammalian cytochrome P450 2B4 | | Descriptor: | Cytochrome P450 2B4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Scott, E.E, He, Y.A, Wester, M.R, White, M.A, Chin, C.C, Halpert, J.R, Johnson, E.F, Stout, C.D. | | Deposit date: | 2003-06-13 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An open conformation of mammalian cytochrome P450 2B4 at 1.6 A resolution
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
2K6U
 
 | | The Solution Structure of a Conformationally Restricted Fully Active Derivative of the Human Relaxin-like Factor (RLF) | | Descriptor: | Insulin-like 3 A chain, Insulin-like 3 B chain | | Authors: | Bullesbach, E.E, Hass, M.A.S, Jensen, M.R, Hansen, D.F, Kristensen, S.M, Schwabe, C, Led, J.J. | | Deposit date: | 2008-07-24 | | Release date: | 2008-12-16 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a conformationally restricted fully active derivative of the human relaxin-like factor
Biochemistry, 47, 2008
|
|
2OHO
 
 | |
2KCE
 
 | |
