4X9Q
 
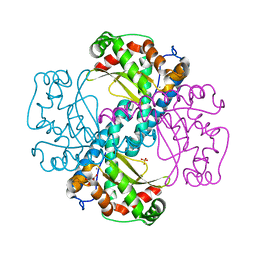 | | MnSOD-3 Room Temperature Structure | | Descriptor: | MALONATE ION, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Hunter, G.J, Trinh, C.H, Hunter, T, Bonetta, R, Stewart, E.E. | | Deposit date: | 2014-12-11 | | Release date: | 2015-11-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The structure of the Caenorhabditis elegans manganese superoxide dismutase MnSOD-3-azide complex.
Protein Sci., 24, 2015
|
|
4NKW
 
 | |
5C8C
 
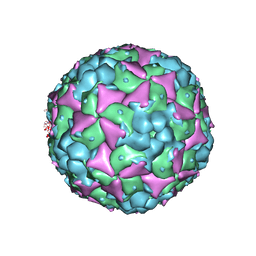 | | Crystal structure of recombinant coxsackievirus A16 capsid | | Descriptor: | CHLORIDE ION, POTASSIUM ION, STEARIC ACID, ... | | Authors: | Ren, J, Wang, X, Zhu, L, Hu, Z, Gao, Q, Yang, P, Li, X, Wang, J, Shen, X, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2015-06-25 | | Release date: | 2015-09-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of Coxsackievirus A16 Capsids with Native Antigenicity: Implications for Particle Expansion, Receptor Binding, and Immunogenicity.
J.Virol., 89, 2015
|
|
5CTT
 
 | |
5CTQ
 
 | |
4OGW
 
 | | Structure of a human CD38 mutant complexed with NMN | | Descriptor: | ADP-ribosyl cyclase 1, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE | | Authors: | Shewchuk, L.M, Preugschat, F, Carter, L.H, Boros, E.E, Moyer, M.B, Stewart, E.L, Porter, D.J.T. | | Deposit date: | 2014-01-16 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A pre-steady state and steady state kinetic analysis of the N-ribosyl hydrolase activity of hCD157.
Arch.Biochem.Biophys., 564C, 2014
|
|
4Z3U
 
 | | PRV nuclear egress complex | | Descriptor: | CHLORIDE ION, SODIUM ION, UL31, ... | | Authors: | Bigalke, J.M, Heldwein, E.E. | | Deposit date: | 2015-03-31 | | Release date: | 2015-11-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.706 Å) | | Cite: | Structural basis of membrane budding by the nuclear egress complex of herpesviruses.
Embo J., 34, 2015
|
|
4NKV
 
 | |
4NKX
 
 | |
4NKY
 
 | |
5ED7
 
 | | Crystal Structure of HSV-1 UL21 C-terminal Domain | | Descriptor: | CHLORIDE ION, Tegument protein UL21 | | Authors: | Metrick, C.M, Heldwein, E.E. | | Deposit date: | 2015-10-20 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Novel Structure and Unexpected RNA-Binding Ability of the C-Terminal Domain of Herpes Simplex Virus 1 Tegument Protein UL21.
J.Virol., 90, 2016
|
|
4NKZ
 
 | | Human steroidogenic cytochrome P450 17A1 mutant A105L with substrate 17alpha-hydroxypregnenolone | | Descriptor: | (3alpha,8alpha)-3,17-dihydroxypregn-5-en-20-one, PROTOPORPHYRIN IX CONTAINING FE, Steroid 17-alpha-hydroxylase/17,20 lyase | | Authors: | Scott, E.E, Petrunak, E.M. | | Deposit date: | 2013-11-13 | | Release date: | 2014-10-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Structures of Human Steroidogenic Cytochrome P450 17A1 with Substrates.
J.Biol.Chem., 289, 2014
|
|
408D
 
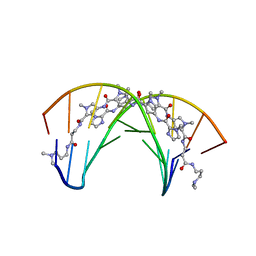 | | STRUCTURAL BASIS FOR RECOGNITION OF A-T AND T-A BASE PAIRS IN THE MINOR GROOVE OF B-DNA | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*TP*AP*CP*TP*GP*G)-3'), IMIDAZOLE-PYRROLE POLYAMIDE | | Authors: | Kielkopf, C.L, White, S, Szewczyk, J.W, Turner, J.M, Baird, E.E, Dervan, P.B, Rees, D.C. | | Deposit date: | 1998-06-24 | | Release date: | 1998-10-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A structural basis for recognition of A.T and T.A base pairs in the minor groove of B-DNA.
Science, 282, 1998
|
|
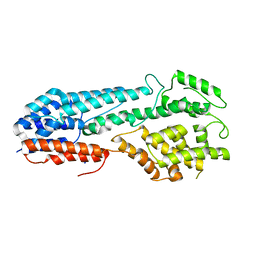
5VYL
 
 | |
5DDJ
 
 | | Crystal structure of recombinant foot-and-mouth-disease virus O1M-S2093Y empty capsid | | Descriptor: | Foot and mouth disease virus, VP1, VP2, ... | | Authors: | Kotecha, A, Seago, J, Scott, K, Burman, A, Loureiro, S, Ren, J, Porta, C, Ginn, H.M, Jackson, T, Perez-Martin, E, Siebert, C.A, Paul, G, Huiskonen, J.T, Jones, I.M, Esnouf, R.M, Fry, E.E, Maree, F.F, Charleston, B, Stuart, D.I. | | Deposit date: | 2015-08-25 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure-based energetics of protein interfaces guides foot-and-mouth disease virus vaccine design.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5D8A
 
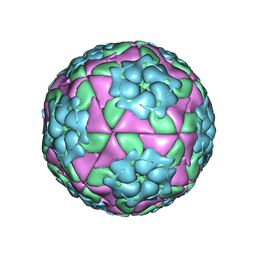 | | Crystal structure of recombinant foot-and-mouth-disease virus A22-H2093F empty capsid | | Descriptor: | VP1, VP2, VP3, ... | | Authors: | Kotecha, A, Seago, J, Scott, K, Burman, A, Loureiro, S, Ren, J, Porta, C, Ginn, H.M, Jackson, T, Perez-Martin, E, Siebert, C.A, Paul, G, Huiskonen, J.T, Jones, I.M, Esnouf, R.M, Fry, E.E, Maree, F.F, Charleston, B, Stuart, D.I. | | Deposit date: | 2015-08-16 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based energetics of protein interfaces guides foot-and-mouth disease virus vaccine design.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5EHU
 
 | |
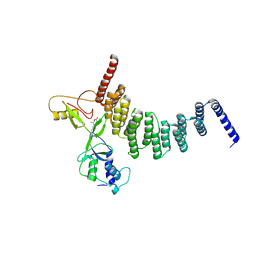
5CTR
 
 | |
5WSN
 
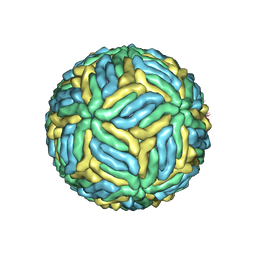 | | Structure of Japanese encephalitis virus | | Descriptor: | E protein, M protein | | Authors: | Wang, X, Zhu, L, Li, S, Yuan, S, Qin, C, Fry, E.E, Stuart, I.D, Rao, Z. | | Deposit date: | 2016-12-07 | | Release date: | 2017-05-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Near-atomic structure of Japanese encephalitis virus reveals critical determinants of virulence and stability
Nat Commun, 8, 2017
|
|
5WTF
 
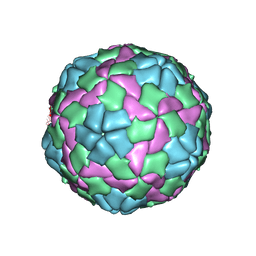 | | Cryo-EM structure for Hepatitis A virus empty particle | | Descriptor: | VP0, VP1, VP3 | | Authors: | Wang, X, Zhu, L, Dang, M, Hu, Z, Gao, Q, Yuan, S, Sun, Y, Zhang, B, Ren, J, Walter, T.S, Wang, J, Fry, E.E, Stuart, D.I, Rao, Z. | | Deposit date: | 2016-12-11 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Potent neutralization of hepatitis A virus reveals a receptor mimic mechanism and the receptor recognition site
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5WTH
 
 | | Cryo-EM structure for Hepatitis A virus complexed with FAB | | Descriptor: | FAB Heavy Chain, FAB Light Chain, Polyprotein, ... | | Authors: | Wang, X, Zhu, L, Dang, M, Hu, Z, Gao, Q, Yuan, S, Sun, Y, Zhang, B, Ren, J, Walter, T.S, Wang, J, Fry, E.E, Stuart, D.I, Rao, Z. | | Deposit date: | 2016-12-12 | | Release date: | 2017-01-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Potent neutralization of hepatitis A virus reveals a receptor mimic mechanism and the receptor recognition site
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5EJJ
 
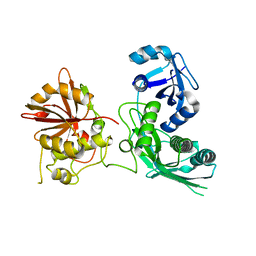 | | Crystal structure of UfSP from C.elegans | | Descriptor: | Ufm1-specific protease | | Authors: | Kim, K, Ha, B, Kim, E.E. | | Deposit date: | 2015-11-02 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The MPN domain of Caenorhabditis elegans UfSP modulates both substrate recognition and deufmylation activity
Biochem. Biophys. Res. Commun., 476, 2016
|
|
5ACA
 
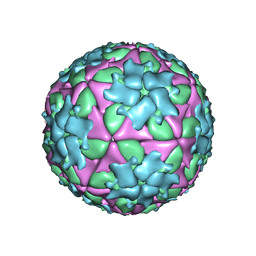 | | Structure-based energetics of protein interfaces guide Foot-and-Mouth disease virus vaccine design | | Descriptor: | VP1, VP2, VP3, ... | | Authors: | Kotecha, A, Seago, J, Scott, K, Burman, A, Loureiro, S, Ren, J, Porta, C, Ginn, H.M, Jackson, T, Perez-Martin, E, Siebert, C.A, Paul, G, Huiskonen, J.T, Jones, I.M, Esnouf, R.M, Fry, E.E, Maree, F.F, Charleston, B, Stuart, D.I. | | Deposit date: | 2015-08-14 | | Release date: | 2015-09-23 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure-Based Energetics of Protein Interfaces Guide Foot-and-Mouth Disease Vaccine Design
Nat.Struct.Mol.Biol., 22, 2015
|
|
5AGT
 
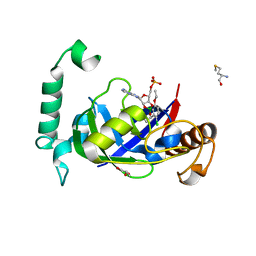 | | Crystal structure of the LeuRS editing domain of Mycobacterium tuberculosis in complex with the adduct (S)-3-(Aminomethyl)-4-chloro-7-ethoxybenzo[c][1,2]oxaborol-1(3H)-ol-AMP | | Descriptor: | 4-Chloro-3-aminomethyl-7-[ethoxy]-3H-benzo[C][1,2]oxaborol-1-ol modified adenosine, GLYCEROL, LEUCINE--TRNA LIGASE, ... | | Authors: | Palencia, A, Li, X, Alley, M.R.K, Ding, C, Easom, E.E, Hernandez, V, Meewan, M, Mohan, M, Rock, F.L, Franzblau, S.G, Wang, Y, Lenaerts, A.J, Parish, T, Cooper, C.B, Waters, M.G, Ma, Z, Mendoza, A, Barros, D, Cusack, S, Plattner, J.J. | | Deposit date: | 2015-02-03 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Discovery of Novel Oral Protein Synthesis Inhibitors of Mycobacterium Tuberculosis that Target Leucyl-tRNA Synthetase.
Antimicrob.Agents Chemother., 60, 2016
|
|
4P5N
 
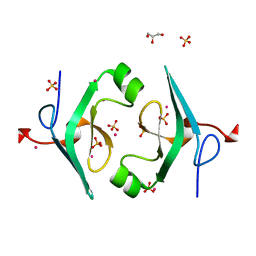 | | Structure of CNAG_02591 from Cryptococcus neoformans | | Descriptor: | CADMIUM ION, GLYCEROL, Hypothetical protein CNAG_02591, ... | | Authors: | Ramagopal, U.A, McClelland, E.E, Toro, R, Casadevall, A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-03-18 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | A Small Protein Associated with Fungal Energy Metabolism Affects the Virulence of Cryptococcus neoformans in Mammals.
Plos Pathog., 12, 2016
|
|
