6MS4
 
 | | Crystal structure of the DENR-MCT-1 complex | | Descriptor: | Density-regulated protein, GLYCEROL, Malignant T-cell-amplified sequence 1, ... | | Authors: | Lomakin, I.B, Steitz, T.A, Dmitriev, S.E. | | Deposit date: | 2018-10-16 | | Release date: | 2019-01-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal structure of the DENR-MCT-1 complex revealed zinc-binding site essential for heterodimer formation.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
8BIB
 
 | | O-Methyltransferase Plu4890 in complex with SAH and AQ-256 | | Descriptor: | 1,3,8-tris(oxidanyl)anthracene-9,10-dione, CARBONATE ION, CHLORIDE ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2022-11-02 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A set of closely related methyltransferases for site-specific tailoring of anthraquinone pigments.
Structure, 31, 2023
|
|
6GM4
 
 | | [FeFe]-hydrogenase CpI from Clostridium pasteurianum, variant S319A | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, IRON/SULFUR CLUSTER, Iron hydrogenase 1, ... | | Authors: | Duan, J, Esselborn, J, Hofmann, E, Winkler, M, Happe, T. | | Deposit date: | 2018-05-24 | | Release date: | 2018-11-07 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystallographic and spectroscopic assignment of the proton transfer pathway in [FeFe]-hydrogenases.
Nat Commun, 9, 2018
|
|
8BIE
 
 | | O-Methyltransferase Plu4894 in complex with SAH | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, methyltransferase Plu4894 | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2022-11-02 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A set of closely related methyltransferases for site-specific tailoring of anthraquinone pigments.
Structure, 31, 2023
|
|
6MMH
 
 | | Diheteromeric NMDA receptor GluN1/GluN2A in the 'Extended-2' conformation, in complex with glycine and glutamate, in the presence of 1 millimolar zinc chloride, and at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, NMDA 1, ... | | Authors: | Jalali-Yazdi, F, Chowdhury, S, Yoshioka, C, Gouaux, E. | | Deposit date: | 2018-09-30 | | Release date: | 2018-11-28 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (8.21 Å) | | Cite: | Mechanisms for Zinc and Proton Inhibition of the GluN1/GluN2A NMDA Receptor.
Cell, 175, 2018
|
|
7S2H
 
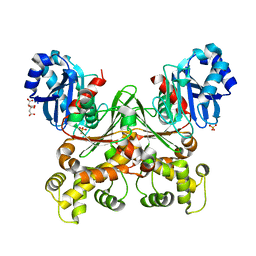 | | Crystal structure of Trypanosoma cruzi glucokinase in the apo form (open conformation) | | Descriptor: | CITRATE ANION, Glucokinase 1, putative, ... | | Authors: | Kearns, S.P, Daneshian, L, Swartz, P.D, Carey, S.M, Chruszcz, M, D'Antonio, E.L. | | Deposit date: | 2021-09-03 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Trypanosoma cruzi glucokinase in the apo form (open conformation)
To Be Published
|
|
5I3I
 
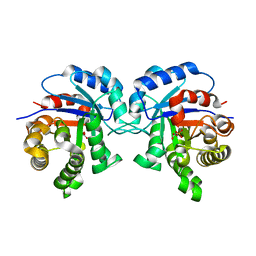 | | Structure-Function Studies on Role of Hydrophobic Clamping of a Basic Glutamate in Catalysis by Triosephosphate Isomerase | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, Triosephosphate isomerase, glycosomal | | Authors: | Drake, E.J, Gulick, A.M, Richard, J.P, Zhai, X, Kim, K, Reinhardt, C.J. | | Deposit date: | 2016-02-10 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Function Studies of Hydrophobic Residues That Clamp a Basic Glutamate Side Chain during Catalysis by Triosephosphate Isomerase.
Biochemistry, 55, 2016
|
|
7S2P
 
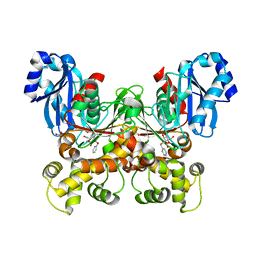 | | Crystal structure of the F337L mutation of Trypanosoma cruzi glucokinase in complex with inhibitor CBZ-GlcN | | Descriptor: | 2-{[(benzyloxy)carbonyl]amino}-2-deoxy-beta-D-glucopyranose, Glucokinase 1, SULFATE ION | | Authors: | Carey, S.M, Nettles, R.B, Daneshian, L, Chruszcz, M, D'Antonio, E.L. | | Deposit date: | 2021-09-03 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of the F337L mutation of Trypanosoma cruzi glucokinase in complex with inhibitor CBZ-GlcN
To Be Published
|
|
9AYY
 
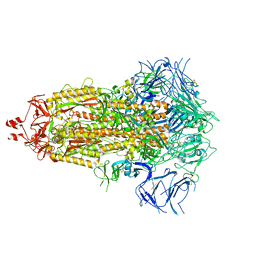 | |
6MUR
 
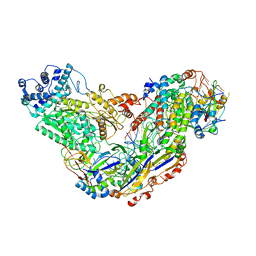 | | Cryo-EM structure of Csm-crRNA-target RNA ternary complex in type III-A CRISPR-Cas system | | Descriptor: | RNA (27-MER), RNA (5'-R(P*GP*CP*AP*AP*AP*CP*CP*GP*CP*CP*UP*CP*UP*GP*CP*CP*CP*GP*C)-3'), Uncharacterized protein, ... | | Authors: | Jia, N, Wang, C, Eng, E.T, Patel, D.J. | | Deposit date: | 2018-10-23 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Type III-A CRISPR-Cas Csm Complexes: Assembly, Periodic RNA Cleavage, DNase Activity Regulation, and Autoimmunity.
Mol. Cell, 73, 2019
|
|
8QRI
 
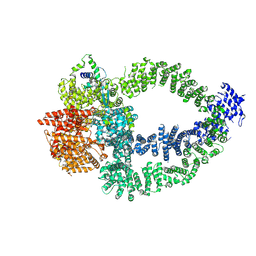 | | TRRAP and EP400 in the human Tip60 complex | | Descriptor: | E1A-binding protein p400, Transformation/transcription domain-associated protein | | Authors: | Li, C, Smirnova, E, Schnitzler, C, Crucifix, C, Concordet, J.P, Brion, A, Poterszman, A, SChultz, P, Papai, G, Ben-Shem, A. | | Deposit date: | 2023-10-09 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the human TIP60-C histone exchange and acetyltransferase complex.
Nature, 2024
|
|
5FOW
 
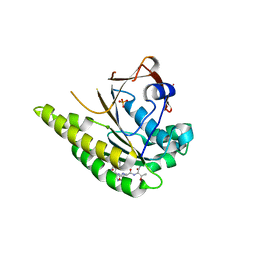 | | HUMANISED MONOMERIC RADA IN COMPLEX WITH WHTA TETRAPEPTIDE | | Descriptor: | DNA repair and recombination protein RadA, PHOSPHATE ION, WHTA PEPTIDE | | Authors: | Scott, D.E, Marsh, M, Blundell, T.L, Abell, C, Hyvonen, M. | | Deposit date: | 2015-11-26 | | Release date: | 2016-03-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | Structure Activity Relationship of the Peptide Binding Motif Mediating the Rad51:Brca2 Protein-Protein Interaction.
FEBS Lett., 590, 2016
|
|
7UEG
 
 | |
4WVJ
 
 | | Crystal structure of the Type-I signal peptidase from Staphylococcus aureus (SpsB) in complex with an inhibitor peptide (pep3). | | Descriptor: | Maltose-binding periplasmic protein,Signal peptidase IB, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, inhibitor peptide (PEP3) | | Authors: | Young, P.G, Ting, Y.T, Baker, E.N. | | Deposit date: | 2014-11-05 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Peptide binding to a bacterial signal peptidase visualized by peptide tethering and carrier-driven crystallization.
IUCrJ, 3, 2016
|
|
6R1B
 
 | | Crystal structure of UgpB from Mycobacterium tuberculosis in complex with glycerophosphocholine | | Descriptor: | 2-(((R)-2,3-DIHYDROXYPROPYL)PHOSPHORYLOXY)-N,N,N-TRIMETHYLETHANAMINIUM, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Fenn, J, Nepravishta, R, Guy, C.S, Harrison, J, Angulo, J, Cameron, A.D, Fullam, E. | | Deposit date: | 2019-03-14 | | Release date: | 2019-09-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.27000213 Å) | | Cite: | Structural Basis of Glycerophosphodiester Recognition by theMycobacterium tuberculosisSubstrate-Binding Protein UgpB.
Acs Chem.Biol., 14, 2019
|
|
5IS0
 
 | | Structure of TRPV1 in complex with capsazepine, determined in lipid nanodisc | | Descriptor: | Transient receptor potential cation channel subfamily V member 1, capsazepine | | Authors: | Gao, Y, Cao, E, Julius, D, Cheng, Y. | | Deposit date: | 2016-03-15 | | Release date: | 2016-05-25 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action.
Nature, 534, 2016
|
|
7UYV
 
 | | Crystal structure of JAK3 kinase domain in complex with compound 25 | | Descriptor: | 6-{[(2M)-2-(2-chloro-6-fluorophenyl)-5-oxo-5H-pyrrolo[3,4-b]pyridin-4-yl]amino}-N-ethylpyridine-3-carboxamide, CHLORIDE ION, Tyrosine-protein kinase JAK3 | | Authors: | Toms, A.V, Leit, S, Greenwood, J.R, Mondal, S, Carriero, S, Dahlgren, M, Harriman, G.C, Kennedy-Smith, J.J, Kapeller, R, Lawson, J.P, Romero, D.L, Shelley, M, Wester, R.T, Westlin, W, Mc Elwee, J.J, Miao, W, Edmondson, S.D, Massee, C.E. | | Deposit date: | 2022-05-07 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Potent and selective TYK2-JH1 inhibitors highly efficacious in rodent model of psoriasis.
Bioorg.Med.Chem.Lett., 73, 2022
|
|
1UNM
 
 | | Crystal structure of 7-Aminoactinomycin D with non-complementary DNA | | Descriptor: | 5'-D(*TP*TP*AP*GP*BRU*TP)-3', 7-AMINOACTINOMYCIN D | | Authors: | Alexopoulos, E.C, Klement, R, Jares-Erijman, E.A, Uson, I, Jovin, T.M, Sheldrick, G.M. | | Deposit date: | 2003-09-11 | | Release date: | 2004-09-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal and Solution Structures of 7-Amino-Actinomycin D Complexes with D(Ttagbrut), D(Ttagtt) and D(Tttagttt)
Acta Crystallogr.,Sect.D, 61, 2005
|
|
7ROY
 
 | | The structure of the Fem1B:FNIP1 complex | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Folliculin-interacting protein 1, Protein fem-1 homolog B, ... | | Authors: | Gee, C.L, Mena, E.L, Manford, A.G, Rape, M. | | Deposit date: | 2021-08-02 | | Release date: | 2021-10-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis and regulation of the reductive stress response.
Cell, 184, 2021
|
|
8B07
 
 | |
6MUT
 
 | | Cryo-EM structure of ternary Csm-crRNA-target RNA with anti-tag sequence complex in type III-A CRISPR-Cas system | | Descriptor: | RNA (5'-R(*GP*UP*GP*GP*AP*AP*AP*GP*GP*CP*GP*GP*GP*CP*AP*GP*AP*GP*GP*C)-3'), RNA (5'-R(P*CP*CP*UP*CP*UP*GP*CP*CP*CP*GP*CP*CP*UP*UP*UP*CP*CP*AP*C)-3'), Uncharacterized protein Csm1, ... | | Authors: | Jia, N, Wang, C, Eng, E.T, Patel, D.J. | | Deposit date: | 2018-10-23 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Type III-A CRISPR-Cas Csm Complexes: Assembly, Periodic RNA Cleavage, DNase Activity Regulation, and Autoimmunity.
Mol. Cell, 73, 2019
|
|
9AYW
 
 | |
6BWH
 
 | |
5I0V
 
 | |
6FHJ
 
 | | Structural dynamics and catalytic properties of a multi-modular xanthanase, native. | | Descriptor: | CALCIUM ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Moroz, O.V, Jensen, P.F, McDonald, S.P, McGregor, N, Blagova, E, Comamala, G, Segura, D.R, Anderson, L, Vasu, S.M, Rao, V.P, Giger, L, Monrad, R.N, Svendsen, A, Nielsen, J.E, Henrissat, B, Davies, G.J, Brumer, H, Rand, K, Wilson, K.S. | | Deposit date: | 2018-01-14 | | Release date: | 2018-08-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural Dynamics and Catalytic Properties of a Multimodular Xanthanase
Acs Catalysis, 2018
|
|
