8CWE
 
 | | 20ns Temperature-Jump (Dark2) XFEL structure of Lysozyme Bound to N,N'-diacetylchitobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
8CVV
 
 | | 20ns Temperature-Jump (Dark1) XFEL structure of Lysozyme | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-18 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
8SAG
 
 | |
8CWD
 
 | | 20ns Temperature-Jump (Dark1) XFEL structure of Lysozyme Bound to N,N'-diacetylchitobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
4XI2
 
 | |
8CVW
 
 | | 20ns Temperature-Jump (Dark2) XFEL structure of Lysozyme | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-18 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
5L7R
 
 | | Crystal structure of BvGH123 | | Descriptor: | 1,2-ETHANEDIOL, glycoside hydrolase | | Authors: | Roth, C, Petricevic, M, John, A, Goddard-Borger, E.D, Davies, G.J, Williams, S.J. | | Deposit date: | 2016-06-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and mechanistic insights into a Bacteroides vulgatus retaining N-acetyl-beta-galactosaminidase that uses neighbouring group participation.
Chem. Commun. (Camb.), 52, 2016
|
|
8CWF
 
 | | 200us Temperature-Jump (Light) XFEL structure of Lysozyme Bound to N,N'-diacetylchitobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
7SJX
 
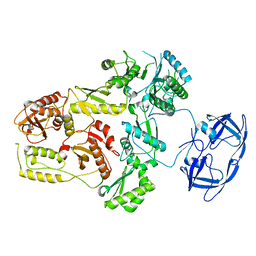 | | Cryo-EM Structure of the PR-RT components of the HIV-1 Pol Polyprotein | | Descriptor: | Gag-Pol polyprotein | | Authors: | Lyumkis, D, Passos, D, Arnold, E, Harrison, J.J.E.K, Ruiz, F.X. | | Deposit date: | 2021-10-19 | | Release date: | 2022-07-27 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Cryo-EM structure of the HIV-1 Pol polyprotein provides insights into virion maturation.
Sci Adv, 8, 2022
|
|
8CU6
 
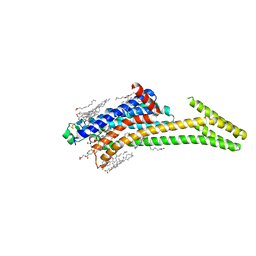 | | Crystal structure of A2AAR-StaR2-S277-bRIL in complex with a novel A2a antagonist, LJ-4517 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2R,3R,4R)-2-[(8P)-6-amino-2-(hex-1-yn-1-yl)-8-(thiophen-2-yl)-9H-purin-9-yl]oxolane-3,4-diol, Adenosine receptor A2a,Soluble cytochrome b562, ... | | Authors: | Shiriaeva, A, Park, D.-J, Kim, G, Lee, Y, Hou, X, Jarhad, D.B, Kim, G, Yu, J, Hyun, Y.E, Kim, W, Gao, Z.-G, Jacobson, K.A, Han, G.W, Stevens, R.C, Jeong, L.S, Choi, S, Cherezov, V. | | Deposit date: | 2022-05-16 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | GPCR Agonist-to-Antagonist Conversion: Enabling the Design of Nucleoside Functional Switches for the A 2A Adenosine Receptor.
J.Med.Chem., 65, 2022
|
|
6DHE
 
 | | RT XFEL structure of the dark-stable state of Photosystem II (0F, S1-rich) at 2.05 Angstrom resolution | | Descriptor: | (6'R,11cis,11'cis,13cis,15cis)-4',5'-didehydro-5',6'-dihydro-beta,beta-carotene, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Kern, J, Chatterjee, R, Young, I.D, Fuller, F.D, Lassalle, L, Ibrahim, M, Gul, S, Fransson, T, Brewster, A.S, Alonso-Mori, R, Hussein, R, Zhang, M, Douthit, L, de Lichtenberg, C, Cheah, M.H, Shevela, D, Wersig, J, Seufert, I, Sokaras, D, Pastor, E, Weninger, C, Kroll, T, Sierra, R.G, Aller, P, Butryn, A, Orville, A.M, Liang, M, Batyuk, A, Koglin, J.E, Carbajo, S, Boutet, S, Moriarty, N.W, Holton, J.M, Dobbek, H, Adams, P.D, Bergmann, U, Sauter, N.K, Zouni, A, Messinger, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2018-05-20 | | Release date: | 2018-11-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structures of the intermediates of Kok's photosynthetic water oxidation clock.
Nature, 563, 2018
|
|
5LBG
 
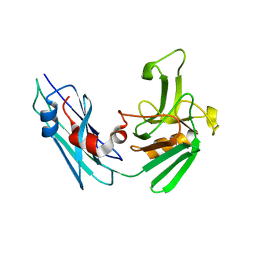 | | Crystal structure of the Mycobacterium tuberculosis L,D-transpeptidase-2 (LdtMt2) BC-module with faropenem-derived adduct at the active site cysteine-354 | | Descriptor: | (3S)-3-HYDROXYBUTANOIC ACID, L,D-transpeptidase 2 | | Authors: | Steiner, E.M, Schnell, R, Schneider, G. | | Deposit date: | 2016-06-16 | | Release date: | 2017-01-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Binding and processing of beta-lactam antibiotics by the transpeptidase LdtMt2 from Mycobacterium tuberculosis.
FEBS J., 284, 2017
|
|
7L3R
 
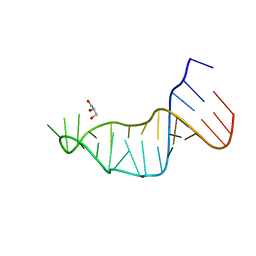 | |
7Q5I
 
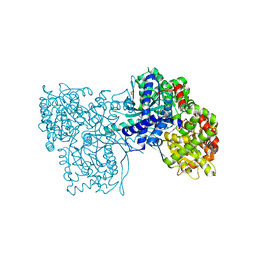 | | A glucose-based molecular rotor probes the catalytic site of glycogen phosphorylase. | | Descriptor: | 2-cyano-3-[4-(dimethylamino)phenyl]-~{N}-[(2~{R},3~{R},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]propanamide, BETA-MERCAPTOETHANOL, CARBONATE ION, ... | | Authors: | Neofytos, D.D, Chrysina, E.D. | | Deposit date: | 2021-11-03 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A glucose-based molecular rotor inhibitor of glycogen phosphorylase as a probe of cellular enzymatic function.
Org.Biomol.Chem., 20, 2022
|
|
8FRA
 
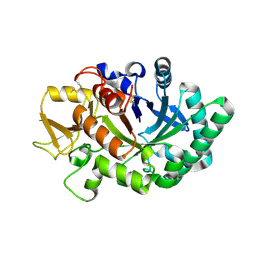 | | Mouse acidic mammalian chitinase, catalytic domain in complex with diacetylchitobiose at pH 5.60 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Acidic mammalian chitinase, ... | | Authors: | Diaz, R.E, Fraser, J.S. | | Deposit date: | 2023-01-06 | | Release date: | 2023-03-08 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural characterization of ligand binding and pH-specific enzymatic activity of mouse Acidic Mammalian Chitinase.
Biorxiv, 2024
|
|
7ZE7
 
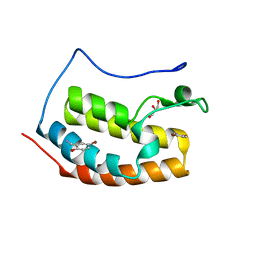 | | BRD4 in complex with FragLite21 | | Descriptor: | 5-bromo-2-hydroxybenzonitrile, Bromodomain-containing protein 4, GLYCEROL | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-03-30 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
8FRC
 
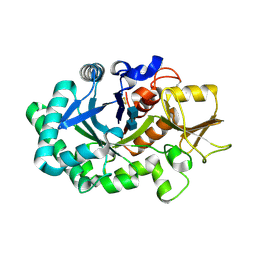 | |
8CU7
 
 | | Crystal structure of A2AAR-StaR2-bRIL in complex with a novel A2a antagonist, LJ-4517 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2R,3R,4R)-2-[(8P)-6-amino-2-(hex-1-yn-1-yl)-8-(thiophen-2-yl)-9H-purin-9-yl]oxolane-3,4-diol, Adenosine receptor A2a,Soluble cytochrome b562, ... | | Authors: | Shiriaeva, A, Park, D.-J, Kim, G, Lee, Y, Hou, X, Jarhad, D.B, Kim, G, Yu, J, Hyun, Y.E, Kim, W, Gao, Z.-G, Jacobson, K.A, Han, G.W, Stevens, R.C, Jeong, L.S, Choi, S, Cherezov, V. | | Deposit date: | 2022-05-16 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | GPCR Agonist-to-Antagonist Conversion: Enabling the Design of Nucleoside Functional Switches for the A 2A Adenosine Receptor.
J.Med.Chem., 65, 2022
|
|
6TCW
 
 | | Crystal structure of Bacteroides thetaiotamicron EndoBT-3987 with Man5GlcNAc product | | Descriptor: | CALCIUM ION, Endo-beta-N-acetylglucosaminidase F1, GLYCEROL, ... | | Authors: | Trastoy, B, Du, J.J, Klontz, E.H, Cifuente, J.O, Sundberg, E.J, Guerin, M.E. | | Deposit date: | 2019-11-06 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Structural basis of mammalian high-mannose N-glycan processing by human gut Bacteroides.
Nat Commun, 11, 2020
|
|
6H6K
 
 | | The structure of the FKR mutant of the archaeal translation initiation factor 2 gamma subunit in complex with GDPCP, obtained in the absence of magnesium salts in the crystallization solution. | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, SODIUM ION, ... | | Authors: | Nikonov, O, Kravchenko, O, Nevskaya, N, Stolboushkina, E, Gabdulkhakov, A, Garber, M, Nikonov, S. | | Deposit date: | 2018-07-27 | | Release date: | 2019-04-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The third structural switch in the archaeal translation initiation factor 2 (aIF2) molecule and its possible role in the initiation of GTP hydrolysis and the removal of aIF2 from the ribosome.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
8VGI
 
 | |
8FRB
 
 | | Mouse acidic mammalian chitinase, catalytic domain in complex with N,N'-diacetylchitobiose at pH 5.25 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetylamino-2-deoxy-alpha-L-idopyranose, ... | | Authors: | Diaz, R.E, Fraser, J.S. | | Deposit date: | 2023-01-06 | | Release date: | 2023-03-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural characterization of ligand binding and pH-specific enzymatic activity of mouse Acidic Mammalian Chitinase.
Biorxiv, 2024
|
|
5L4J
 
 | | Crystal Structure of Human Transthyretin in Complex with 4,4'-Dihydroxydiphenyl sulfone (Bisphenol S, BPS) | | Descriptor: | 4-(4-hydroxyphenyl)sulfonylphenol, SODIUM ION, Transthyretin | | Authors: | Grundstrom, C, Hall, M, Zhang, J, Olofsson, A, Andersson, P, Sauer-Eriksson, A.E. | | Deposit date: | 2016-05-25 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure-Based Virtual Screening Protocol for in Silico Identification of Potential Thyroid Disrupting Chemicals Targeting Transthyretin.
Environ. Sci. Technol., 50, 2016
|
|
6DHO
 
 | | RT XFEL structure of the two-flash state of Photosystem II (2F, S3-rich) at 2.07 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Chatterjee, R, Young, I.D, Fuller, F.D, Lassalle, L, Ibrahim, M, Gul, S, Fransson, T, Brewster, A.S, Alonso-Mori, R, Hussein, R, Zhang, M, Douthit, L, de Lichtenberg, C, Cheah, M.H, Shevela, D, Wersig, J, Seufert, I, Sokaras, D, Pastor, E, Weninger, C, Kroll, T, Sierra, R.G, Aller, P, Butryn, A, Orville, A.M, Liang, M, Batyuk, A, Koglin, J.E, Carbajo, S, Boutet, S, Moriarty, N.W, Holton, J.M, Dobbek, H, Adams, P.D, Bergmann, U, Sauter, N.K, Zouni, A, Messinger, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2018-05-20 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structures of the intermediates of Kok's photosynthetic water oxidation clock.
Nature, 563, 2018
|
|
8FRD
 
 | |
