4XKT
 
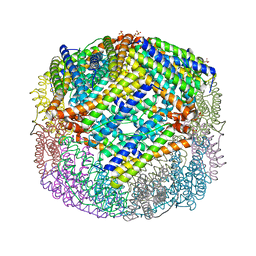 | | E coli BFR variant Y149F | | Descriptor: | Bacterioferritin, SULFATE ION | | Authors: | Bradley, J.M, Hemmings, A.M, Le Brun, N.E. | | Deposit date: | 2015-01-12 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Three Aromatic Residues are Required for Electron Transfer during Iron Mineralization in Bacterioferritin.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
8P67
 
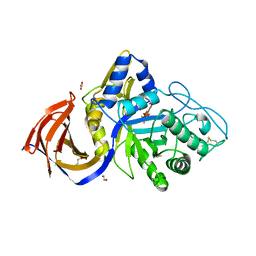 | | Crystal structure of Thermothelomyces thermophila (double mutant EE) in complex with aldotetrauronic acid | | Descriptor: | 1,2-ETHANEDIOL, 4-O-methyl-alpha-D-glucopyranuronic acid-(1-2)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, GH30 family xylanase, ... | | Authors: | Dimarogona, M, Pentari, C, Kosinas, C, Topakas, E. | | Deposit date: | 2023-05-25 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structural and molecular insights into a bifunctional glycoside hydrolase 30 xylanase specific to glucuronoxylan.
Biotechnol.Bioeng., 121, 2024
|
|
2MHQ
 
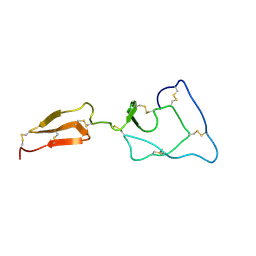 | | Solution structure of the major factor VIII binding region on von Willebrand factor | | Descriptor: | von Willebrand factor | | Authors: | Shiltagh, N, Kirkpatrick, J, Cabrita, L.D, McKinnon, T.A.J, Thalassinos, K, Tuddenham, E.G.D, Hansen, D.F. | | Deposit date: | 2013-12-02 | | Release date: | 2014-05-14 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the major factor VIII binding region on von Willebrand factor.
Blood, 123, 2014
|
|
4XAV
 
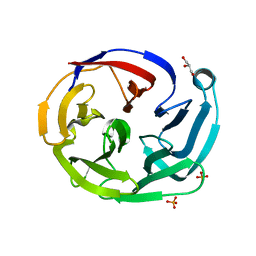 | | Crystal structure of olfactomedin domain from gliomedin | | Descriptor: | GLYCEROL, Gliomedin, PHOSPHATE ION, ... | | Authors: | Hill, S.E, Nguyen, E, Lieberman, R.L. | | Deposit date: | 2014-12-15 | | Release date: | 2015-07-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Molecular Details of Olfactomedin Domains Provide Pathway to Structure-Function Studies.
Plos One, 10, 2015
|
|
8T4O
 
 | | Human mitochondrial serine hydroxymethyltransferase (SHMT2) in complex with PLP, glycine and AGF347 inhibitor with no glutamate | | Descriptor: | 4-[4-(2-amino-4-oxo-3,4-dihydro-5H-pyrrolo[3,2-d]pyrimidin-5-yl)butyl]-2-fluorobenzoic acid, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], Serine hydroxymethyltransferase, ... | | Authors: | Katinas, J.M, Dann III, C.E. | | Deposit date: | 2023-06-09 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structural Characterization of 5-Substituted Pyrrolo[3,2- d ]pyrimidine Antifolate Inhibitors in Complex with Human Serine Hydroxymethyl Transferase 2.
Biochemistry, 2024
|
|
6AVB
 
 | | CryoEM structure of Mical Oxidized Actin (Class 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle | | Authors: | Grintsevich, E.E, Ge, P, Sawaya, M.R, Yesilyurt, H.G, Terman, J.R, Zhou, Z.H, Reisler, E. | | Deposit date: | 2017-09-01 | | Release date: | 2018-01-17 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Catastrophic disassembly of actin filaments via Mical-mediated oxidation.
Nat Commun, 8, 2017
|
|
6E7L
 
 | | Structure of a dimerized UUCG motif | | Descriptor: | RNA (5'-R(*GP*GP*GP*CP*UP*GP*CP*AP*CP*UP*UP*CP*GP*GP*UP*GP*CP*UP*GP*CP*CP*C)-3') | | Authors: | Montemayor, E.J, Butcher, S.E. | | Deposit date: | 2018-07-26 | | Release date: | 2019-07-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structure of an RNA helix with pyrimidine mismatches and cross-strand stacking.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6M3F
 
 | |
2IH4
 
 | |
7PGE
 
 | | copper transporter PcoB | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Copper resistance protein B, LAURYL DIMETHYLAMINE-N-OXIDE, ... | | Authors: | Li, P, Gourdon, P.E. | | Deposit date: | 2021-08-13 | | Release date: | 2022-07-06 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | PcoB is a defense outer membrane protein that facilitates cellular uptake of copper.
Protein Sci., 31, 2022
|
|
8TL6
 
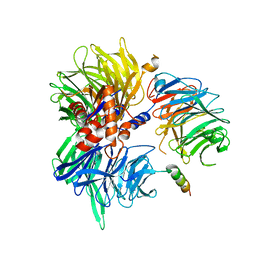 | | Cryo-EM structure of DDB1deltaB-DDA1-DCAF5 | | Descriptor: | DDB1- and CUL4-associated factor 5, DET1- and DDB1-associated protein 1, DNA damage-binding protein 1 | | Authors: | Yue, H, Hunkeler, M, Roy Burman, S.S, Fischer, E.S. | | Deposit date: | 2023-07-26 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | Targeting DCAF5 suppresses SMARCB1-mutant cancer by stabilizing SWI/SNF.
Nature, 628, 2024
|
|
4PUO
 
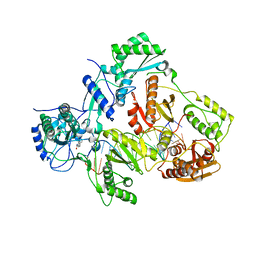 | | Crystal structure of HIV-1 reverse transcriptase in complex with RNA/DNA and Nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, 5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*G)-3', 5'-R(P*AP*UP*GP*GP*UP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*UP*GP*UP*G)-3', ... | | Authors: | Das, K, Martinez, S.E, Arnold, E. | | Deposit date: | 2014-03-13 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Structures of HIV-1 RT-RNA/DNA ternary complexes with dATP and nevirapine reveal conformational flexibility of RNA/DNA: insights into requirements for RNase H cleavage.
Nucleic Acids Res., 42, 2014
|
|
1S1X
 
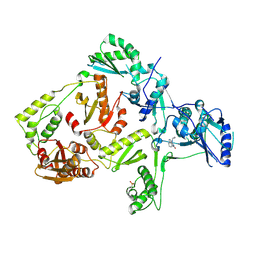 | | Crystal structure of V108I mutant HIV-1 reverse transcriptase in complex with nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, Reverse transcriptase | | Authors: | Ren, J, Nichols, C.E, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2004-01-07 | | Release date: | 2004-06-29 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of HIV-1 reverse transcriptases mutated at codons 100, 106 and 108 and mechanisms of resistance to non-nucleoside inhibitors
J.Mol.Biol., 336, 2004
|
|
8TLU
 
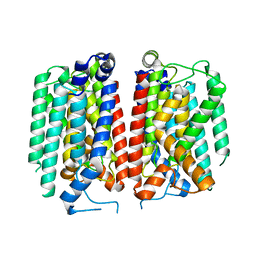 | | E. coli MraY mutant-T23P | | Descriptor: | Phospho-N-acetylmuramoyl-pentapeptide-transferase | | Authors: | Orta, A.K, Li, Y.E, Clemons, W.M. | | Deposit date: | 2023-07-27 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Synthesis of lipid-linked precursors of the bacterial cell wall is governed by a feedback control mechanism in Pseudomonas aeruginosa.
Nat Microbiol, 9, 2024
|
|
4XAT
 
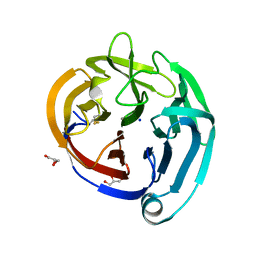 | | Crystal structure of the olfactomedin domain from noelin/pancortin/olfactomedin-1 | | Descriptor: | CALCIUM ION, GLYCEROL, Noelin, ... | | Authors: | Hill, S.E, Nguyen, E, Lieberman, R.L. | | Deposit date: | 2014-12-15 | | Release date: | 2015-07-15 | | Last modified: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (2.113 Å) | | Cite: | Molecular Details of Olfactomedin Domains Provide Pathway to Structure-Function Studies.
Plos One, 10, 2015
|
|
1B0P
 
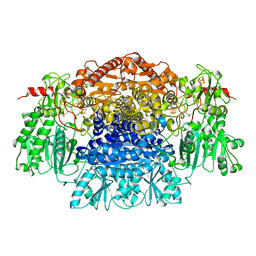 | | CRYSTAL STRUCTURE OF PYRUVATE-FERREDOXIN OXIDOREDUCTASE FROM DESULFOVIBRIO AFRICANUS | | Descriptor: | CALCIUM ION, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Chabriere, E, Charon, M.H, Volbeda, A. | | Deposit date: | 1998-11-12 | | Release date: | 1999-04-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal structures of the key anaerobic enzyme pyruvate:ferredoxin oxidoreductase, free and in complex with pyruvate.
Nat.Struct.Biol., 6, 1999
|
|
8SXZ
 
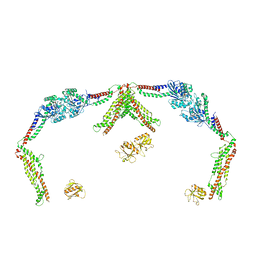 | |
8IPZ
 
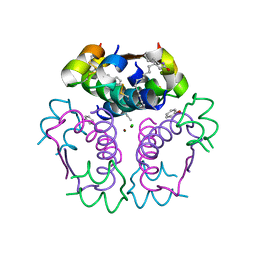 | |
8T0R
 
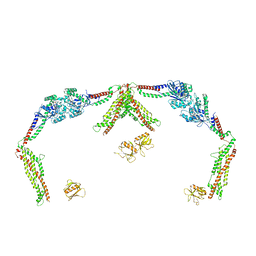 | |
7Z6U
 
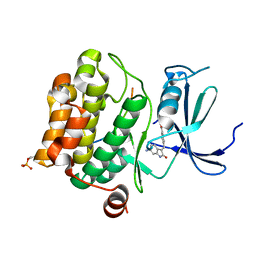 | | Pim1 in complex with (E)-4-((6-amino-2-oxoindolin-3-ylidene)methyl)benzoic acid and Pimtide | | Descriptor: | 4-[(~{E})-(6-azanyl-2-oxidanylidene-1~{H}-indol-3-ylidene)methyl]benzoic acid, GLYCEROL, Isoform 1 of Serine/threonine-protein kinase pim-1, ... | | Authors: | Hochban, P.M.M, Heine, A, Diederich, W.E. | | Deposit date: | 2022-03-14 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Pose, duplicate, then elaborate: Steps towards increased affinity for inhibitors targeting the specificity surface of the Pim-1 kinase.
Eur.J.Med.Chem., 245, 2023
|
|
6MCY
 
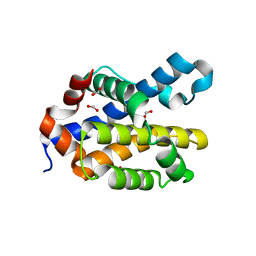 | | Crystal structure of mouse Bak | | Descriptor: | 1,2-ETHANEDIOL, Bcl-2 homologous antagonist/killer, FORMIC ACID | | Authors: | Brouwer, J.M, Czabotar, P.E, Colman, P.M. | | Deposit date: | 2018-09-03 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.748 Å) | | Cite: | A small molecule interacts with VDAC2 to block mouse BAK-driven apoptosis.
Nat.Chem.Biol., 15, 2019
|
|
8SZ8
 
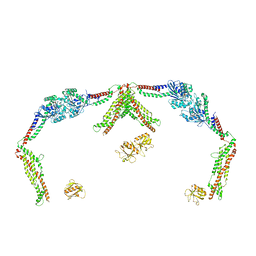 | |
8T0K
 
 | |
8PRN
 
 | | E1M, K50A, R52A, D97A, E99A MUTANT OF RH. BLASTICA PORIN | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, PORIN | | Authors: | Maveyraud, L, Schmid, B, Schulz, G.E. | | Deposit date: | 1998-06-12 | | Release date: | 1998-08-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Porin mutants with new channel properties.
Protein Sci., 7, 1998
|
|
7PV2
 
 | | GA1 bacteriophage portal protein | | Descriptor: | Head-tail connector (Portal protein) | | Authors: | Javed, A, Villanueva, H, Orlova, E.V, Savva, R. | | Deposit date: | 2021-10-01 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM Structures of Two Bacteriophage Portal Proteins Provide Insights for Antimicrobial Phage Engineering.
Viruses, 13, 2021
|
|
