6XOD
 
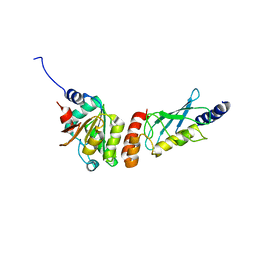 | | Crystal structure of the PEX4-PEX22 protein complex from Arabidopsis thaliana | | Descriptor: | Peroxisome biogenesis protein 22, Protein PEROXIN-4 | | Authors: | Olmos Jr, J.L, Bradford, S.E, Miller, M.D, Xu, W, Wright, Z.J, Bartel, B, Phillips Jr, G.N. | | Deposit date: | 2020-07-06 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The Structure of the Arabidopsis PEX4-PEX22 Peroxin Complex-Insights Into Ubiquitination at the Peroxisomal Membrane
Front Cell Dev Biol, 10, 2022
|
|
6EN5
 
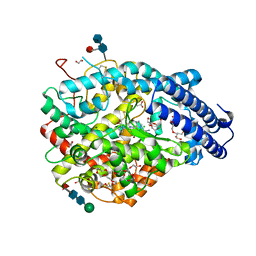 | | Crystal structure A of the Angiotensin-1 converting enzyme N-domain in complex with a diprolyl inhibitor. | | Descriptor: | (2~{S})-1-[(2~{S})-2-[[(1~{S})-1-[(2~{S})-1-[(2~{S})-2-azanyl-4-oxidanyl-4-oxidanylidene-butanoyl]pyrrolidin-2-yl]-2-oxidanyl-2-oxidanylidene-ethyl]amino]propanoyl]pyrrolidine-2-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cozier, G.E, Acharya, K.R, Fienberg, S, Chibale, K, Sturrock, E.D. | | Deposit date: | 2017-10-04 | | Release date: | 2017-12-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Design and Development of a Potent and Selective Novel Diprolyl Derivative That Binds to the N-Domain of Angiotensin-I Converting Enzyme.
J. Med. Chem., 61, 2018
|
|
2ICY
 
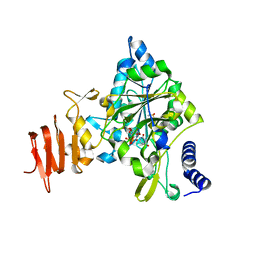 | | Crystal Structure of a Putative UDP-glucose Pyrophosphorylase from Arabidopsis Thaliana with Bound UDP-glucose | | Descriptor: | DIMETHYL SULFOXIDE, Probable UTP-glucose-1-phosphate uridylyltransferase 2, URIDINE-5'-DIPHOSPHATE-GLUCOSE, ... | | Authors: | McCoy, J.G, Wesenberg, G.E, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-09-13 | | Release date: | 2006-10-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure and Dynamics of UDP-Glucose Pyrophosphorylase from Arabidopsis thaliana with Bound UDP-Glucose and UTP.
J.Mol.Biol., 366, 2007
|
|
6U2M
 
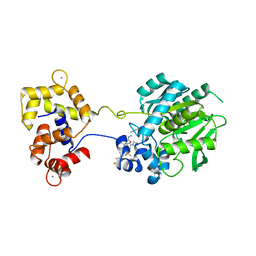 | | Crystal structure of a HaloTag-based calcium indicator, HaloCaMP V2, bound to JF635 | | Descriptor: | (1E,3S)-1-{10-[2-carboxy-5-({2-[2-(hexyloxy)ethoxy]ethyl}carbamoyl)phenyl]-7-(3-fluoroazetidin-1-yl)-5,5-dimethyldibenz o[b,e]silin-3(5H)-ylidene}-3-fluoroazetidin-1-ium, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Deo, C, Schreiter, E.R. | | Deposit date: | 2019-08-20 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The HaloTag as a general scaffold for far-red tunable chemigenetic indicators.
Nat.Chem.Biol., 17, 2021
|
|
2IDO
 
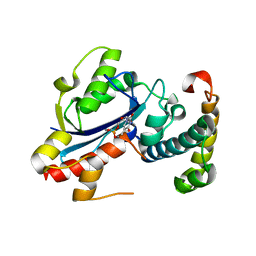 | | Structure of the E. coli Pol III epsilon-Hot proofreading complex | | Descriptor: | 1,2-ETHANEDIOL, DNA polymerase III epsilon subunit, Hot protein, ... | | Authors: | Kirby, T.W, Harvey, S, DeRose, E.F, Chalov, S, Chikova, A.K, Perrino, F.W, Schaaper, R.M, London, R.E, Pedersen, L.C. | | Deposit date: | 2006-09-15 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the Escherichia coli DNA polymerase III epsilon-HOT proofreading complex.
J.Biol.Chem., 281, 2006
|
|
6ZTG
 
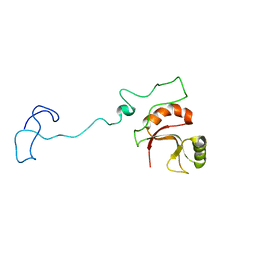 | | Spor protein DedD | | Descriptor: | Cell division protein DedD | | Authors: | Pazos, M, Peters, K, Boes, A, Safaei, Y, Kenward, C, Caveney, N.A, Laguri, C, Breukink, E, Strynadka, N.C.J, Simorre, J.P, Terrak, M, Vollmer, W. | | Deposit date: | 2020-07-20 | | Release date: | 2020-11-11 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | SPOR Proteins Are Required for Functionality of Class A Penicillin-Binding Proteins in Escherichia coli.
Mbio, 11, 2020
|
|
6S71
 
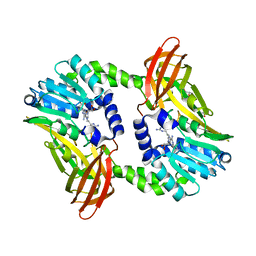 | | Crystal structure of CARM1 in complex with inhibitor WH5C | | Descriptor: | (2~{S})-4-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-(5-carbamimidamidopentyl)amino]-2-azanyl-butanoic acid, GLYCEROL, Histone-arginine methyltransferase CARM1 | | Authors: | Gunnell, E.A, Muhsen, U, Dowden, J, Dreveny, I. | | Deposit date: | 2019-07-04 | | Release date: | 2020-03-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.062 Å) | | Cite: | Structural and biochemical evaluation of bisubstrate inhibitors of protein arginine N-methyltransferases PRMT1 and CARM1 (PRMT4).
Biochem.J., 477, 2020
|
|
6S7B
 
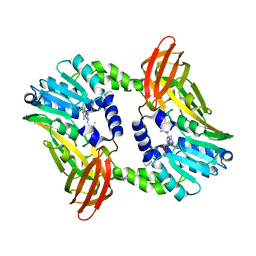 | | Crystal structure of CARM1 in complex with inhibitor UM249 | | Descriptor: | 1-[4-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-(3-azanylpropyl)amino]butyl]guanidine, Histone-arginine methyltransferase CARM1 | | Authors: | Gunnell, E.A, Muhsen, U, Dowden, J, Dreveny, I. | | Deposit date: | 2019-07-04 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.659 Å) | | Cite: | Structural and biochemical evaluation of bisubstrate inhibitors of protein arginine N-methyltransferases PRMT1 and CARM1 (PRMT4).
Biochem.J., 477, 2020
|
|
6ARB
 
 | | CRYSTAL STRUCTURE OF PROTEIN CitE FROM MYCOBACTERIUM TUBERCULOSIS IN COMPLEX WITH MAGNESIUM, PYRUVATE AND COENZYME A | | Descriptor: | COENZYME A, Citrate lyase subunit beta-like protein, GLYCEROL, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Wang, H, Bonanno, J.B, Carvalho, L, Almo, S.C. | | Deposit date: | 2017-08-22 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.717 Å) | | Cite: | An essential bifunctional enzyme inMycobacterium tuberculosisfor itaconate dissimilation and leucine catabolism.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6RS0
 
 | | GOLGI ALPHA-MANNOSIDASE II in complex with (2S,3S,4R,5R)-1-(2-(Benzyloxy)ethyl)-2-(hydroxymethyl)piperidine-3,4,5-triol | | Descriptor: | (2~{S},3~{S},4~{R})-2-(hydroxymethyl)-1-(2-phenylmethoxyethyl)piperidine-3,4,5-triol, 1,2-ETHANEDIOL, Alpha-mannosidase 2, ... | | Authors: | Armstrong, Z, Lahav, D, Johnson, R, Kuo, C.L, Beenakker, T.J.M, de Boer, C, Wong, C.S, van Rijssel, E.R, Debets, M, Geurink, P.P, Ovaa, H, van der Stelt, M, Codee, J.D.C, Aerts, J.M.F.G, Wu, L, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2019-05-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Manno- epi -cyclophellitols Enable Activity-Based Protein Profiling of Human alpha-Mannosidases and Discovery of New Golgi Mannosidase II Inhibitors.
J.Am.Chem.Soc., 142, 2020
|
|
4R7W
 
 | | Crystal structure of 5-methylcytosine deaminase from Klebsiella pneumoniae liganded with phosphonocytosine | | Descriptor: | (2R)-2-amino-2,5-dihydro-1,5,2-diazaphosphinin-6(1H)-one 2-oxide, 1,2-ETHANEDIOL, Cytosine deaminase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Hitchcock, D.S, Raushel, F.M, Almo, S.C. | | Deposit date: | 2014-08-28 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Crystal structure of 5-methylcytosine deaminase from Klebsiella pneumoniae liganded with phosphonocytosine
To be Published
|
|
5YAK
 
 | |
6UJX
 
 | | HIV-1 wild-type reverse transcriptase-DNA complex with (-)-FTC-TP | | Descriptor: | DNA primer, DNA template, MAGNESIUM ION, ... | | Authors: | Lansdon, E.B. | | Deposit date: | 2019-10-03 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.70451212 Å) | | Cite: | Elucidating molecular interactions ofL-nucleotides with HIV-1 reverse transcriptase and mechanism of M184V-caused drug resistance.
Commun Biol, 2, 2019
|
|
2ITF
 
 | | Crystal structure IsdA NEAT domain from Staphylococcus aureus with heme bound | | Descriptor: | Iron-regulated surface determinant protein A, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Grigg, J.C, Vermeiren, C.L, Heinrichs, D.E, Murphy, M.E. | | Deposit date: | 2006-10-19 | | Release date: | 2006-12-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Haem recognition by a Staphylococcus aureus NEAT domain.
Mol.Microbiol., 63, 2007
|
|
6XKO
 
 | |
5C84
 
 | | Fragment-Based Drug Discovery Targeting Inhibitor of Apoptosis Proteins: Compound 20 | | Descriptor: | (2R,5R)-4-[2-(6-chloro-3,3-dimethyl-2,3-dihydro-1H-pyrrolo[3,2-c]pyridin-1-yl)-2-oxoethyl]-5-(methoxymethyl)-2-methylpiperazin-1-ium, E3 ubiquitin-protein ligase XIAP, ZINC ION | | Authors: | Chessari, G, Buck, I.M, Day, J.E.H, Day, P.J, Iqbal, A, Johnson, C.N, Lewis, E.J, Martins, V, Miller, D, Reader, M, Rees, D.C, Rich, S.J, Tamanini, E, Vitorino, M, Ward, G.A, Williams, P.A, Williams, G, Wilsher, N.E, Woolford, A.J.-A. | | Deposit date: | 2015-06-25 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Fragment-Based Drug Discovery Targeting Inhibitor of Apoptosis Proteins: Discovery of a Non-Alanine Lead Series with Dual Activity Against cIAP1 and XIAP.
J.Med.Chem., 58, 2015
|
|
6EWO
 
 | | Crystal structure of non-phosphorylated form of RTF PHOSPHOPEPTIDE BOUND TO HLA-A2 in complex with LILRB1 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Mohammed, F, Stones, D.H, Willcox, B.E. | | Deposit date: | 2017-11-06 | | Release date: | 2018-11-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Application of the immunoregulatory receptor LILRB1 as a crystallisation chaperone for human class I MHC complexes.
J. Immunol. Methods, 464, 2019
|
|
6RMX
 
 | | Human Carbonic anhydrase II mutant T199V bound by 2-[(1S)-2,3-Dihydro-1H-inden-1-ylamino]-3,5,6-trifluoro-4-[(2-hydroxyethyl)thio]benzenesulfonamide | | Descriptor: | 2-[(1S)-2,3-dihydro-1H-inden-1-ylamino]-3,5,6-trifluoro-4-[(2-hydroxyethyl)sulfanyl]benzenesulfonamide, BENZOIC ACID, Carbonic anhydrase 2, ... | | Authors: | Smirnov, A, Paketuryte, V, Manakova, E, Grazulis, S. | | Deposit date: | 2019-05-07 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Human Carbonic anhydrase II mutant T199V bound by 2-[(1S)-2,3-Dihydro-1H-inden-1-ylamino]-3,5,6-trifluoro-4-[(2-hydroxyethyl)thio]benzenesulfonamide
To Be Published
|
|
6EXM
 
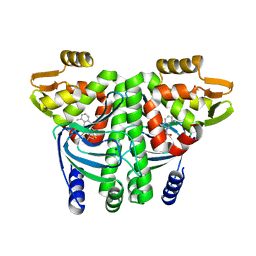 | | The Transcriptional Regulator PrfA from Listeria Monocytogenes in complex with a ring-fused 2-pyridone (MK202) | | Descriptor: | ISOPROPYL ALCOHOL, Listeriolysin positive regulatory factor A, SODIUM ION, ... | | Authors: | Grundstrom, C, Begum, A, Hall, M, Kulen, M, Lindgren, M, Johansson, J, Almqvist, F, Sauer, U.H, Sauer-Eriksson, A.E. | | Deposit date: | 2017-11-08 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Based Design of Inhibitors Targeting PrfA, the Master Virulence Regulator of Listeria monocytogenes.
J. Med. Chem., 61, 2018
|
|
6RVU
 
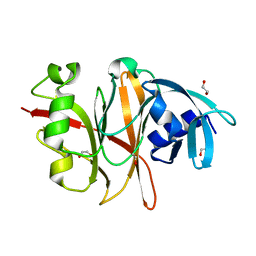 | | Crystal structure of the Burkholderia Lethal Factor 1 (BLF1) | | Descriptor: | 1,2-ETHANEDIOL, Lethal Factor 1 (BLF1) | | Authors: | Mobbs, G.W, Aziz, A.A, Blackburn, G.M, Sedelnikova, S.E, Minshull, T.C, Dickman, M.J, Baker, P.J, Nathan, S, Firdaus-Raih, M, Rice, D.W. | | Deposit date: | 2019-06-01 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Molecular basis of specificity and deamidation of eIF4A by Burkholderia Lethal Factor 1.
Commun Biol, 5, 2022
|
|
6EY2
 
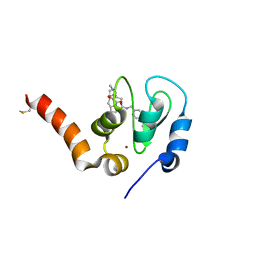 | | Crystal structure of XIAP-BIR3 in complex with a cIAP1-selective SM | | Descriptor: | (3~{S},6~{S},7~{S},9~{a}~{S})-~{N}-[(4-~{tert}-butylphenyl)methyl]-7-(hydroxymethyl)-6-[[(2~{S})-2-(methylamino)butanoyl]amino]-5-oxidanylidene-1,2,3,6,7,8,9,9~{a}-octahydropyrrolo[1,2-a]azepine-3-carboxamide, E3 ubiquitin-protein ligase XIAP, ZINC ION | | Authors: | Cossu, F, Corti, A, Milani, M, Mastrangelo, E. | | Deposit date: | 2017-11-10 | | Release date: | 2018-08-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based design and molecular profiling of Smac-mimetics selective for cellular IAPs.
FEBS J., 285, 2018
|
|
6I23
 
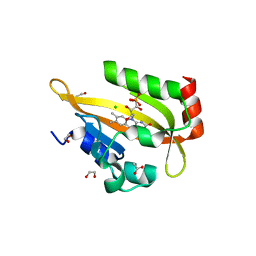 | | Flavin Analogue Sheds Light on Light-Oxygen-Voltage Domain Mechanism | | Descriptor: | 1,2-ETHANEDIOL, 1-deoxy-1-(7,8-dimethyl-2,4-dioxo-3,4-dihydropyrimido[4,5-b]quinolin-10(2H)-yl)-5-O-phosphono-D-ribitol, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Rizkallah, P.J, Kalvaitis, M.E, Allemann, R.K, Mart, R.J, Johnson, L.A. | | Deposit date: | 2018-10-31 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Noncanonical Chromophore Reveals Structural Rearrangements of the Light-Oxygen-Voltage Domain upon Photoactivation.
Biochemistry, 58, 2019
|
|
6RPA
 
 | | Crystal structure of the T-cell receptor NYE_S2 bound to HLA A2*01-SLLMWITQV | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Coles, C.H, Mulvaney, R, Malla, S, Lloyd, A, Smith, K, Chester, F, Knox, A, Stacey, A.R, Dukes, J, Baston, E, Griffin, S, Vuidepot, A, Jakobsen, B.K, Harper, S. | | Deposit date: | 2019-05-14 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | TCRs with Distinct Specificity Profiles Use Different Binding Modes to Engage an Identical Peptide-HLA Complex.
J Immunol., 204, 2020
|
|
6RXA
 
 | | EDDS lyase variant D290M/Y320M with bound formate | | Descriptor: | Argininosuccinate lyase, FORMIC ACID, GLYCEROL, ... | | Authors: | Grandi, E, Poelarends, G.J, Thunnissen, A.M.W.H. | | Deposit date: | 2019-06-07 | | Release date: | 2019-10-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Engineered C-N Lyase: Enantioselective Synthesis of Chiral Synthons for Artificial Dipeptide Sweeteners.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
5IUA
 
 | | Crystal structure of stabilized A2A adenosine receptor A2AR-StaR2-bRIL in complex with compound 12b at 2.2A resolution | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-(furan-2-yl)-N~5~-[3-(4-phenylpiperazin-1-yl)propyl][1,2,4]triazolo[1,5-a][1,3,5]triazine-5,7-diamine, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, ... | | Authors: | Segala, E, Guo, D, Cheng, R.K.Y, Bortolato, A, Deflorian, F, Dore, A.S, Errey, J.C, Heitman, L.H, Ijzerman, A.P, Marshall, F.H, Cooke, R.M. | | Deposit date: | 2016-03-17 | | Release date: | 2016-06-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Controlling the Dissociation of Ligands from the Adenosine A2A Receptor through Modulation of Salt Bridge Strength.
J.Med.Chem., 59, 2016
|
|
