2VTH
 
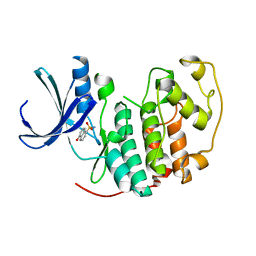 | | Identification of N-(4-piperidinyl)-4-(2,6-dichlorobenzoylamino)-1H-pyrazole-3-carboxamide (AT7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design | | Descriptor: | 5-hydroxynaphthalene-1-sulfonamide, CELL DIVISION PROTEIN KINASE 2, GLYCEROL | | Authors: | Wyatt, P.G, Woodhead, A.J, Boulstridge, J.A, Berdini, V, Carr, M.G, Cross, D.M, Danillon, D, Davis, D.J, Devine, L.A, Early, T.R, Feltell, R.E, Lewis, E.J, McMenamin, R.L, Navarro, E.F, O'Brien, M.A, O'Reilly, M, Reule, M, Saxty, G, Seavers, L.C.A, Smith, D, Squires, M.S, Trewartha, G, Walker, M.T, Woolford, A.J. | | Deposit date: | 2008-05-15 | | Release date: | 2008-08-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of N-(4-Piperidinyl)-4-(2,6-Dichlorobenzoylamino)-1H-Pyrazole-3-Carboxamide (at7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design.
J.Med.Chem., 51, 2008
|
|
5IRX
 
 | | Structure of TRPV1 in complex with DkTx and RTX, determined in lipid nanodisc | | Descriptor: | (2S)-2-(acetyloxy)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}propyl pentanoate, (2S)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(hexanoyloxy)propyl hexanoate, (4R,7S)-4-hydroxy-N,N,N-trimethyl-4,9-dioxo-7-[(pentanoyloxy)methyl]-3,5,8-trioxa-4lambda~5~-phosphatetradecan-1-aminium, ... | | Authors: | Gao, Y, Cao, E, Julius, D, Cheng, Y. | | Deposit date: | 2016-03-14 | | Release date: | 2016-05-25 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action.
Nature, 534, 2016
|
|
2VTP
 
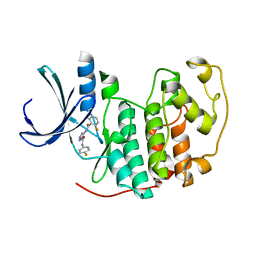 | | Identification of N-(4-piperidinyl)-4-(2,6-dichlorobenzoylamino)-1H- pyrazole-3-carboxamide (AT7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design. | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, {[(2,6-difluorophenyl)carbonyl]amino}-N-(4-fluorophenyl)-1H-pyrazole-3-carboxamide | | Authors: | Wyatt, P.G, Woodhead, A.J, Boulstridge, J.A, Berdini, V, Carr, M.G, Cross, D.M, Danillon, D, Davis, D.J, Devine, L.A, Early, T.R, Feltell, R.E, Lewis, E.J, McMenamin, R.L, Navarro, E.F, O'Brien, M.A, O'Reilly, M, Reule, M, Saxty, G, Seavers, L.C.A, Smith, D, Squires, M.S, Trewartha, G, Walker, M.T, Woolford, A.J. | | Deposit date: | 2008-05-15 | | Release date: | 2008-08-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Identification of N-(4-Piperidinyl)-4-(2,6-Dichlorobenzoylamino)-1H-Pyrazole-3-Carboxamide (at7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design.
J.Med.Chem., 51, 2008
|
|
7ZV9
 
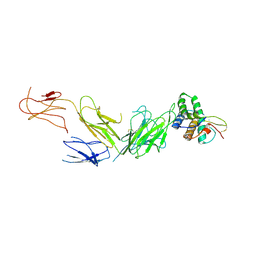 | |
1L6X
 
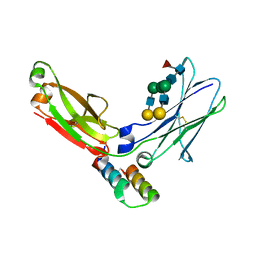 | | FC FRAGMENT OF RITUXIMAB BOUND TO A MINIMIZED VERSION OF THE B-DOMAIN FROM PROTEIN A CALLED Z34C | | Descriptor: | IMMUNOGLOBULIN GAMMA-1 HEAVY CHAIN CONSTANT REGION, Minimized B-domain of Protein A Z34C, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Idusogie, E.E, Presta, L.G, Santoro-Gazzano, H, Totpal, K, Wong, P.Y, Ultsch, M, Meng, Y.G, Mullkerrin, M.G. | | Deposit date: | 2002-03-14 | | Release date: | 2002-04-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mapping of the C1q binding site on rituxan, a chimeric antibody with a human IgG1 Fc.
J.Immunol., 164, 2000
|
|
2IF7
 
 | | Crystal Structure of NTB-A | | Descriptor: | CALCIUM ION, CHLORIDE ION, SLAM family member 6 | | Authors: | Cao, E, Ramagopal, U.A, Fedorov, A.A, Fedorov, E.V, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2006-09-20 | | Release date: | 2006-10-17 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | NTB-A Receptor Crystal Structure: Insights into Homophilic Interactions in the Signaling Lymphocytic Activation Molecule Receptor Family.
Immunity, 25, 2006
|
|
6NEN
 
 | | Catalytic domain of Proteus mirabilis ScsC | | Descriptor: | Copper resistance protein | | Authors: | Kurth, F, Furlong, E.J, Premkumar, L, Martin, J.L. | | Deposit date: | 2018-12-17 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Engineered variants provide new insight into the structural properties important for activity of the highly dynamic, trimeric protein disulfide isomerase ScsC from Proteus mirabilis.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
2IN6
 
 | | Wee1 kinase complex with inhibitor PD311839 | | Descriptor: | 3-(9-HYDROXY-1,3-DIOXO-4-PHENYL-2,3-DIHYDROPYRROLO[3,4-C]CARBAZOL-6(1H)-YL)PROPANOIC ACID, Wee1-like protein kinase | | Authors: | Squire, C.J, Dickson, J.M, Ivanovic, I, Baker, E.N. | | Deposit date: | 2006-10-05 | | Release date: | 2007-09-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthesis and structure-activity relationships of N-6 substituted analogues of 9-hydroxy-4-phenylpyrrolo[3,4-c]carbazole-1,3(2H,6H)-diones as inhibitors of Wee1 and Chk1 checkpoint kinases.
Eur.J.Med.Chem., 43, 2008
|
|
1A4W
 
 | | CRYSTAL STRUCTURES OF THROMBIN WITH THIAZOLE-CONTAINING INHIBITORS: PROBES OF THE S1' BINDING SITE | | Descriptor: | ALPHA-THROMBIN (LARGE SUBUNIT), ALPHA-THROMBIN (SMALL SUBUNIT), HIRUGEN, ... | | Authors: | Matthews, J.H, Krishnan, R, Costanzo, M.J, Maryanoff, B.E, Tulinsky, A. | | Deposit date: | 1998-02-06 | | Release date: | 1998-04-29 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of thrombin with thiazole-containing inhibitors: probes of the S1' binding site.
Biophys.J., 71, 1996
|
|
5CDF
 
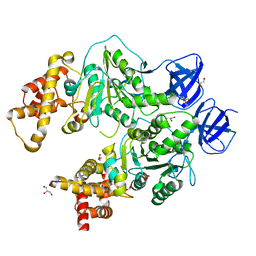 | | Structure at 2.3 A of the alpha/beta monomer of the F-ATPase from Paracoccus denitrificans | | Descriptor: | ATP synthase subunit alpha, ATP synthase subunit beta, GLYCEROL, ... | | Authors: | Morales-Rios, E, Montgomery, M.G, Leslie, A.G.W, Garcia-Trejo, J.J, Walker, J.E. | | Deposit date: | 2015-07-03 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a catalytic dimer of the alpha- and beta-subunits of the F-ATPase from Paracoccus denitrificans at 2.3 angstrom resolution.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
6NJL
 
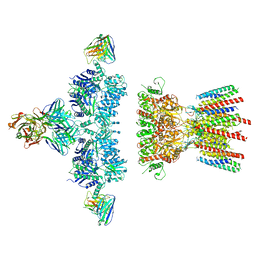 | |
6JL5
 
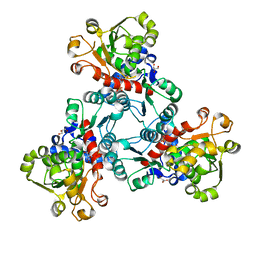 | | Crystal structure of aspartate transcarbamoylase from Trypanosoma cruzi in complex with aspartate (Asp) and phosphate (Pi). | | Descriptor: | ASPARTIC ACID, Aspartate carbamoyltransferase, GLYCEROL, ... | | Authors: | Matoba, K, Shiba, T, Nara, T, Aoki, T, Nagasaki, S, Hayamizu, R, Honma, T, Tanaka, A, Inoue, M, Matsuoka, S, Balogun, E.O, Inaoka, D.K, Kita, K, Harada, S. | | Deposit date: | 2019-03-04 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystallographic snapshots of Trypanosoma cruzi aspartate transcarbamoylase
revealed an ordered Bi-Bi reaction mechanism
To Be Published
|
|
1A46
 
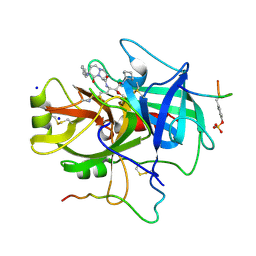 | | THROMBIN COMPLEXED WITH HIRUGEN AND A BETA-STRAND MIMETIC INHIBITOR | | Descriptor: | (1S,7S)-7-amino-N-[(2R,3S)-7-amino-1-(cyclohexylamino)-2-hydroxy-1-oxoheptan-3-yl]-7-benzyl-8-oxohexahydro-1H-pyrazolo[1,2-a]pyridazine-1-carboxamide, ALPHA-THROMBIN (LARGE SUBUNIT), ALPHA-THROMBIN (SMALL SUBUNIT), ... | | Authors: | St Charles, R, Matthews, J.H, Zhang, E, Tulinsky, A, Kahn, M. | | Deposit date: | 1998-02-11 | | Release date: | 1998-05-27 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Bound structures of novel P3-P1' beta-strand mimetic inhibitors of thrombin.
J.Med.Chem., 42, 1999
|
|
1AI3
 
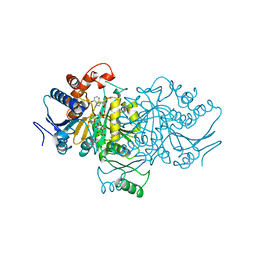 | | ORBITAL STEERING IN THE CATALYTIC POWER OF ENZYMES: SMALL STRUCTURAL CHANGES WITH LARGE CATALYTIC CONSEQUENCES | | Descriptor: | ISOCITRATE DEHYDROGENASE, ISOCITRIC ACID, MAGNESIUM ION, ... | | Authors: | Stoddard, B.L, Mesecar, A, Koshland Junior, D.E. | | Deposit date: | 1997-04-30 | | Release date: | 1997-11-12 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Orbital steering in the catalytic power of enzymes: small structural changes with large catalytic consequences.
Science, 277, 1997
|
|
6W9Y
 
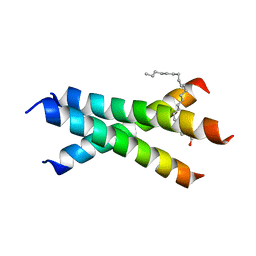 | | De novo designed receptor transmembrane domains enhance CAR-T cytotoxicity and attenuate cytokine release | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, De novo designed receptor transmembrane domain proMP 1.2 | | Authors: | Call, M.J, Call, M.E, Chandler, N.J, Nguyen, J.V, Trenker, R. | | Deposit date: | 2020-03-24 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | De novo designed receptor transmembrane domains enhance CAR-T cytotoxicity and attenuate cytokine release
To Be Published
|
|
4DV2
 
 | | Crystal structure of the Thermus thermophilus 30S ribosomal subunit with a 16S rRNA mutation, C912A | | Descriptor: | 16S rRNA, MAGNESIUM ION, ZINC ION, ... | | Authors: | Demirci, H, Murphy IV, F, Murphy, E, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2012-02-22 | | Release date: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (3.646 Å) | | Cite: | A structural basis for streptomycin resistance
To be Published
|
|
2WAA
 
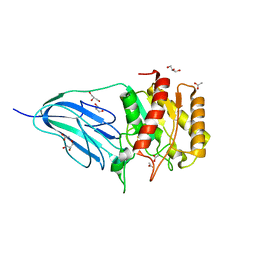 | | Structure of a family two carbohydrate esterase from Cellvibrio japonicus | | Descriptor: | ACETATE ION, GLYCEROL, XYLAN ESTERASE, ... | | Authors: | Montainer, C, Money, V.A, Pires, V.M.R, Flint, J.E, Pinheiro, B.A, Goyal, A, Prates, J.A.M, Izumi, A, Stalbrand, H, Kolenova, K, Topakas, E, Dodson, E.J, Bolam, D.N, Davies, G.J, Fontes, C.M.G.A, Gilbert, H.J. | | Deposit date: | 2009-02-04 | | Release date: | 2009-03-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Active Site of a Carbohydrate Esterase Displays Divergent Catalytic and Noncatalytic Binding Functions.
Plos Biol., 7, 2009
|
|
1TGV
 
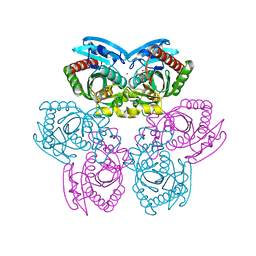 | | Structure of E. coli Uridine Phosphorylase complexed with 5-Fluorouridine and sulfate | | Descriptor: | 5-FLUOROURIDINE, POTASSIUM ION, SULFATE ION, ... | | Authors: | Bu, W, Settembre, E.C, Sanders, J.M, Begley, T.P, Ealick, S.E. | | Deposit date: | 2004-05-31 | | Release date: | 2005-06-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of E. coli Uridine Phosphorylase
To be Published, 2004
|
|
6ZN8
 
 | | Crystal structure of the H. influenzae VapXD toxin-antitoxin complex | | Descriptor: | Endoribonuclease VapD, VapX | | Authors: | Bertelsen, M.B, Senissar, M, Nielsen, M.H, Bisiak, F, Cunha, M.V, Molinaro, A.L, Daines, D.A, Brodersen, D.E. | | Deposit date: | 2020-07-06 | | Release date: | 2020-11-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.211 Å) | | Cite: | Structural Basis for Toxin Inhibition in the VapXD Toxin-Antitoxin System.
Structure, 29, 2021
|
|
6NOG
 
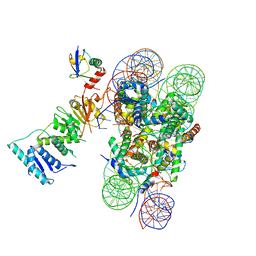 | | Poised-state Dot1L bound to the H2B-Ubiquitinated nucleosome | | Descriptor: | 601 DNA Strand 1, 601 DNA Strand 2, Histone H2A type 1, ... | | Authors: | Worden, E.J, Hoffmann, N.A, Wolberger, C. | | Deposit date: | 2019-01-16 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Mechanism of Cross-talk between H2B Ubiquitination and H3 Methylation by Dot1L.
Cell, 176, 2019
|
|
6NP9
 
 | | PD-L1 IgV domain V76T with fragment | | Descriptor: | Programmed cell death 1 ligand 1, SULFATE ION | | Authors: | Zhao, B, Perry, E. | | Deposit date: | 2019-01-17 | | Release date: | 2019-02-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Fragment-based screening of programmed death ligand 1 (PD-L1).
Bioorg. Med. Chem. Lett., 29, 2019
|
|
6TN4
 
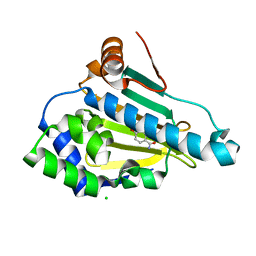 | | Rapid optimisation of fragments and hits to lead compounds from screening of crude reaction mixtures | | Descriptor: | 2,4-bis(oxidanyl)benzamide, CHLORIDE ION, Heat shock protein HSP 90-alpha | | Authors: | Baker, L.M, Aimon, A, Murray, J.B, Surgenor, A.E, Matassova, N, Roughley, S.D, von Delft, F, Hubbard, R.E. | | Deposit date: | 2019-12-05 | | Release date: | 2020-10-14 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.274 Å) | | Cite: | Rapid optimisation of fragments and hits to lead compounds from screening of crude reaction mixtures
Commun Chem, 2020
|
|
6HQP
 
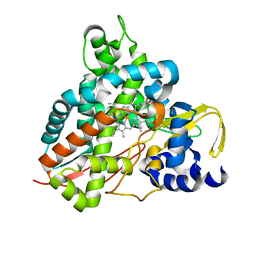 | | Crystal structure of GcoA F169V bound to guaiacol | | Descriptor: | Cytochrome P450, Guaiacol, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mallinson, S.J.B, Hinchen, D.J, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2018-09-25 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Enabling microbial syringol conversion through structure-guided protein engineering.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1KKQ
 
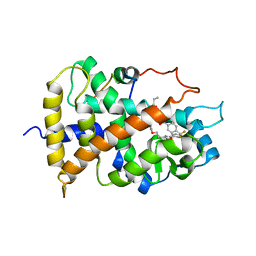 | | Crystal structure of the human PPAR-alpha ligand-binding domain in complex with an antagonist GW6471 and a SMRT corepressor motif | | Descriptor: | N-((2S)-2-({(1Z)-1-METHYL-3-OXO-3-[4-(TRIFLUOROMETHYL) PHENYL]PROP-1-ENYL}AMINO)-3-{4-[2-(5-METHYL-2-PHENYL-1,3-OXAZOL-4-YL)ETHOXY]PHENYL}PROPYL)PROPANAMIDE, NUCLEAR RECEPTOR CO-REPRESSOR 2, PEROXISOME PROLIFERATOR ACTIVATED RECEPTOR | | Authors: | Xu, H.E, Stanley, T.B, Montana, V.G, Lambert, M.H, Shearer, B.G, Cobb, J.E, McKee, D.D, Galardi, C.M, Nolte, R.T, Parks, D.J. | | Deposit date: | 2001-12-10 | | Release date: | 2002-02-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for antagonist-mediated recruitment of nuclear co-repressors by PPARalpha.
Nature, 415, 2002
|
|
8A2E
 
 | | Crystal Structure of Human Parechovirus 3 2A protein | | Descriptor: | 2A protein, GLYCEROL, SULFATE ION | | Authors: | von Castelmur, E, Zhu, L, wang, X, Fry, E, Ren, J, Perrakis, A, Stuart, D.I. | | Deposit date: | 2022-06-03 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural plasticity of 2A proteins in the Parechovirus family.
To Be Published
|
|
