6GXD
 
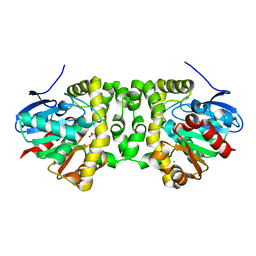 | | The hit-and-return system enables efficient time-resolved serial synchrotron crystallography: FAcD752MS after reaction initiation | | Descriptor: | CHLORIDE ION, fluoroacetate dehalogenase, fluoroacetic acid | | Authors: | Schulz, E.C, Mehrabi, P, Mueller-Werkmeister, H, Tellkamp, F, Stuart, W, Persch, E, De Gasparo, R, Diederich, F, Pai, E.F, Miller, R.J.D. | | Deposit date: | 2018-06-27 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The hit-and-return system enables efficient time-resolved serial synchrotron crystallography.
Nat. Methods, 15, 2018
|
|
6X2N
 
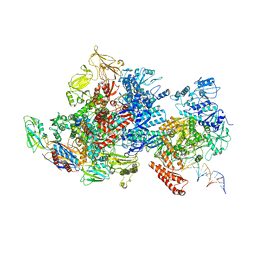 | | Mfd-bound E.coli RNA polymerase elongation complex - I state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (64-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Llewellyn, E, Chen, J, Kang, J.Y, Darst, S.A. | | Deposit date: | 2020-05-20 | | Release date: | 2021-02-03 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for transcription complex disruption by the Mfd translocase.
Elife, 10, 2021
|
|
6ZQ4
 
 | |
7A5V
 
 | | CryoEM structure of a human gamma-aminobutyric acid receptor, the GABA(A)R-beta3 homopentamer, in complex with histamine and megabody Mb25 in lipid nanodisc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Nakane, T, Kotecha, A, Sente, A, Yamashita, K, McMullan, G, Masiulis, S, Brown, P.M.G.E, Grigoras, I.T, Malinauskaite, L, Malinauskas, T, Miehling, J, Yu, L, Karia, D, Pechnikova, E.V, de Jong, E, Keizer, J, Bischoff, M, McCormack, J, Tiemeijer, P, Hardwick, S.W, Chirgadze, D.Y, Murshudov, G, Aricescu, A.R, Scheres, S.H.W. | | Deposit date: | 2020-08-22 | | Release date: | 2020-11-18 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (1.7 Å) | | Cite: | Single-particle cryo-EM at atomic resolution.
Nature, 587, 2020
|
|
8CE9
 
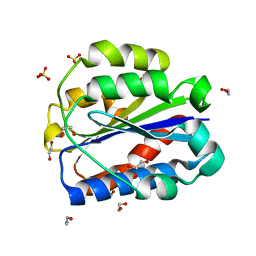 | | Crystal structure of human Cd11b I domain in C121 space group | | Descriptor: | 1,2-ETHANEDIOL, Integrin alpha-M, SULFATE ION | | Authors: | Koekemoer, L, Williams, E, Ni, X, Gao, Q, Coker, J, MacLean, E.M, Wright, N.D, Marsden, B, Von Delft, F, Schwenzer, A, Midwood, K. | | Deposit date: | 2023-02-01 | | Release date: | 2023-03-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal structure of human Cd11b I domain in C121 space group
To Be Published
|
|
7AAA
 
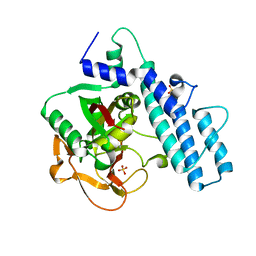 | | Crystal structure of the catalytic domain of human PARP1 (apo) | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Poly [ADP-ribose] polymerase 1, ... | | Authors: | Schimpl, M, Ogden, T.E.H, Yang, J.-C, Underwood, E, Rawlins, P.B, Johannes, J.W, Easton, L.E, Embrey, K.J, Neuhaus, D. | | Deposit date: | 2020-09-04 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Dynamics of the HD regulatory subdomain of PARP-1; substrate access and allostery in PARP activation and inhibition.
Nucleic Acids Res., 49, 2021
|
|
7OFG
 
 | |
8CE6
 
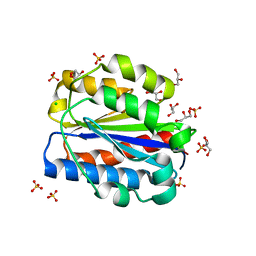 | | Crystal structure of human Cd11b I domain in P212121 space group | | Descriptor: | CHLORIDE ION, GLYCEROL, Integrin alpha-M, ... | | Authors: | Koekemoer, L, Williams, E, Ni, X, Gao, Q, Coker, J, MacLean, E.M, Wright, N.D, Marsden, B, Von Delft, F, Schwenzer, A, Midwood, K. | | Deposit date: | 2023-02-01 | | Release date: | 2023-03-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.582 Å) | | Cite: | Crystal structure of human Cd11b I domain in P212121 space group
To Be Published
|
|
7PAZ
 
 | | REDUCED MUTANT P80I PSEUDOAZURIN FROM A. FAECALIS | | Descriptor: | COPPER (II) ION, PSEUDOAZURIN | | Authors: | Adman, E.T, Libeu, C.A.P. | | Deposit date: | 1997-02-21 | | Release date: | 1997-08-20 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Site-directed mutants of pseudoazurin: explanation of increased redox potentials from X-ray structures and from calculation of redox potential differences.
Biochemistry, 36, 1997
|
|
6GXH
 
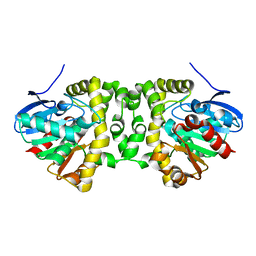 | | The hit-and-return system enables efficient time-resolved serial synchrotron crystallography: FAcD 0MS after reaction initiation | | Descriptor: | Fluoroacetate dehalogenase | | Authors: | Schulz, E.C, Mehrabi, P, Mueller-Werkmeister, H, Tellkamp, F, Stuart, W, Persch, E, De Gasparo, R, Diederich, F, Pai, E.F, Miller, R.J.D. | | Deposit date: | 2018-06-27 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | The hit-and-return system enables efficient time-resolved serial synchrotron crystallography.
Nat. Methods, 15, 2018
|
|
6QDV
 
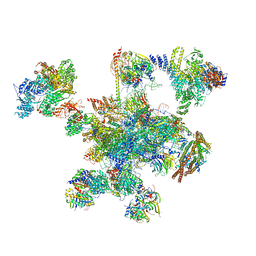 | | Human post-catalytic P complex spliceosome | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent RNA helicase DHX8, ... | | Authors: | Fica, S.M, Oubridge, C, Wilkinson, M.E, Newman, A.J, Nagai, K. | | Deposit date: | 2019-01-03 | | Release date: | 2019-02-20 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A human postcatalytic spliceosome structure reveals essential roles of metazoan factors for exon ligation.
Science, 363, 2019
|
|
7PEL
 
 | | CryoEM structure of simian T-cell lymphotropic virus intasome in complex with PP2A regulatory subunit B56 gamma | | Descriptor: | DNA (5'-D(*AP*CP*TP*GP*TP*GP*TP*TP*TP*GP*GP*CP*GP*CP*TP*TP*CP*TP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*AP*GP*AP*AP*GP*CP*GP*CP*CP*AP*AP*AP*CP*AP*CP*A)-3'), Isoform 3 of PC4 and SFRS1-interacting protein,Isoform Gamma-2 of Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit gamma isoform, ... | | Authors: | Barski, M, Pye, V.E, Nans, A, Cherepanov, P, Maertens, G.N. | | Deposit date: | 2021-08-10 | | Release date: | 2021-08-25 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Cryo-EM structure of the deltaretroviral intasome in complex with the PP2A regulatory subunit B56gamma.
Nat Commun, 11, 2020
|
|
8BO6
 
 | | COAGULATION FACTOR XI PROTEASE DOMAIN IN COMPLEX WITH ACTIVE SITE INHIBITOR 2 | | Descriptor: | (~{E})-~{N}-[[5-(3-azanyl-1~{H}-indazol-6-yl)-4-chloranyl-1~{H}-imidazol-2-yl]methyl]-3-[5-chloranyl-2-(1,2,3,4-tetrazol-1-yl)phenyl]prop-2-enamide, CITRIC ACID, Coagulation factor XIa light chain, ... | | Authors: | Schaefer, M, Roehrig, S, Ackerstaff, J, Nunez, E.J, Gericke, K.M, Meier, K, Tersteegen, A, Stampfuss, J, Ellerbrock, P, Meibom, D, Lang, D, Heitmeier, S, Hillisch, A. | | Deposit date: | 2022-11-14 | | Release date: | 2023-09-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Design and Preclinical Characterization Program toward Asundexian (BAY 2433334), an Oral Factor XIa Inhibitor for the Prevention and Treatment of Thromboembolic Disorders.
J.Med.Chem., 66, 2023
|
|
6GYN
 
 | | Structure of human HCN4 hyperpolarization-activated cyclic nucleotide-gated ion channel | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 | | Authors: | Shintre, C.A, Pike, A.C.W, Tessitore, A, Young, M, Bushell, S.R, Strain-Damerell, C, Mukhopadhyay, S, Burgess-Brown, N.A, Huiskonen, J.T, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-06-30 | | Release date: | 2019-05-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of human HCN4 hyperpolarization-activated cyclic nucleotide-gated ion channel
To Be Published
|
|
6GUA
 
 | | Xylulose 5-phosphate phosphoketolase from Lactococcus lactis | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, Probable phosphoketolase, ... | | Authors: | Scheidig, A.J, Szedlacsek, S.E. | | Deposit date: | 2018-06-19 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-17 | | Method: | SOLUTION SCATTERING (1.95 Å), X-RAY DIFFRACTION | | Cite: | Crystal structure of a xylulose 5-phosphate phosphoketolase. Insights into the substrate specificity for xylulose 5-phosphate.
J.Struct.Biol., 207, 2019
|
|
6NJU
 
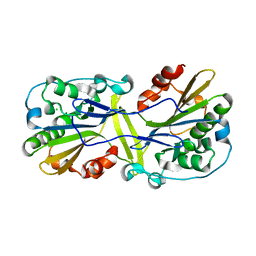 | | Mouse endonuclease G mutant H97A bound to A-DNA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DNA (5'-D(CCGGCGCCGG)-3'), ... | | Authors: | Vander Zanden, C.M, Ho, E.N, Czarny, R.S, Robertson, A.B, Ho, P.S. | | Deposit date: | 2019-01-04 | | Release date: | 2020-01-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural adaptation of vertebrate endonuclease G for 5-hydroxymethylcytosine recognition and function.
Nucleic Acids Res., 48, 2020
|
|
8D8S
 
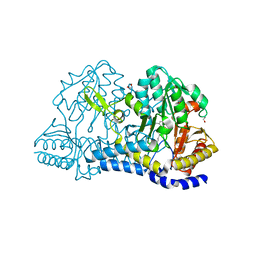 | | SufS from Staphylococcus aureus | | Descriptor: | 1,2-ETHANEDIOL, Cysteine desulfurase | | Authors: | Morrison, C.N, Boncella, A.E, Lunin, V. | | Deposit date: | 2022-06-08 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.388 Å) | | Cite: | Structural and Biochemical Characterization of Staphylococcus aureus Cysteine Desulfurase Complex SufSU.
Acs Omega, 7, 2022
|
|
7P3Y
 
 | |
7PNJ
 
 | |
4YJN
 
 | |
7P3X
 
 | |
7PNR
 
 | | Human Angiogenin mutant-S28AT36AS37A | | Descriptor: | Angiogenin | | Authors: | Papaioannou, O.S.E, Leonidas, D.D. | | Deposit date: | 2021-09-07 | | Release date: | 2021-09-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Biochemical Characterization of the Human Angiogenin-Proliferating Cell Nuclear Antigen Interaction.
Biochemistry, 62, 2023
|
|
6GQT
 
 | | KRAS-169 Q61H GPPNHP + PPIN-2 | | Descriptor: | CITRIC ACID, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Cruz-Migoni, A, Canning, P, Quevedo, C.E, Carr, S.B, Phillips, S.E.V, Rabbitts, T.H. | | Deposit date: | 2018-06-08 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structure-based development of new RAS-effector inhibitors from a combination of active and inactive RAS-binding compounds.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
7P3Z
 
 | |
8CZG
 
 | | Human BAK in complex with the dF3 peptide | | Descriptor: | Bcl-2 homologous antagonist/killer, dF3 peptide | | Authors: | Aguilar, F, Keating, A.E. | | Deposit date: | 2022-05-24 | | Release date: | 2023-01-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Peptides from human BNIP5 and PXT1 and non-native binders of pro-apoptotic BAK can directly activate or inhibit BAK-mediated membrane permeabilization.
Structure, 31, 2023
|
|
