5QDK
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOPL000069a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1, [4-(1H-benzimidazol-1-yl)phenyl]methanol | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.555 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
5QE2
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOPL000398a | | Descriptor: | 1-methyl-N-{[(2S)-oxolan-2-yl]methyl}-1H-pyrazole-3-carboxamide, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.787 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
5QEJ
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMSOA001247b | | Descriptor: | 2,6-dichloropyridine-4-carboxylic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.924 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
5QEZ
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_XST00000713b | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-[(trifluoromethyl)sulfanyl]benzamide, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.654 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
5QFS
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOMB000293a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,4,6,7-tetrahydroacridine-1,8(2H,5H)-dione, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.853 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
5QGB
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOOA000628a | | Descriptor: | (4R,4aR,6R,8aR)-1-benzyloctahydro-2H-6,4-(epiminomethano)-3,1-benzoxazin-2-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.552 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
6ELC
 
 | | Crystal Structure of O-linked Glycosylated VSG3 | | Descriptor: | Variant surface glycoprotein, alpha-D-glucopyranose, alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Stebbins, C.E. | | Deposit date: | 2017-09-28 | | Release date: | 2018-07-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | African trypanosomes evade immune clearance by O-glycosylation of the VSG surface coat.
Nat Microbiol, 3, 2018
|
|
6YNU
 
 | | CaM-P458 complex (crystal form 1) | | Descriptor: | Bifunctional adenylate cyclase toxin/hemolysin CyaA, CALCIUM ION, Calmodulin-1 | | Authors: | Mechaly, A.E, Voegele, A, Haouz, A, Chenal, A. | | Deposit date: | 2020-04-14 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | A High-Affinity Calmodulin-Binding Site in the CyaA Toxin Translocation Domain is Essential for Invasion of Eukaryotic Cells.
Adv Sci, 8, 2021
|
|
6YRF
 
 | | Vip3Bc1 tetramer | | Descriptor: | Vegetative insecticidal protein | | Authors: | Thompson, R.F, Byrne, M.J, Iadanza, M.I, Arribas Perez, M, Maskell, D.P, George, R.M, Hesketh, E.L, Beales, P.A, Zack, M.D, Berry, C. | | Deposit date: | 2020-04-20 | | Release date: | 2021-03-17 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures of an insecticidal Bt toxin reveal its mechanism of action on the membrane.
Nat Commun, 12, 2021
|
|
6ECY
 
 | |
6Y99
 
 | | hSTING mutant R232K in complex with 2',3'-cGAMP | | Descriptor: | Stimulator of interferon genes protein, cGAMP | | Authors: | Boura, E, Smola, M. | | Deposit date: | 2020-03-06 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.984 Å) | | Cite: | Ligand Strain and Its Conformational Complexity Is a Major Factor in the Binding of Cyclic Dinucleotides to STING Protein.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6YDB
 
 | | Human wtSTING in complex with 2',2'-difluoro-3',3'-c-di-GMP | | Descriptor: | 2-azanyl-9-[(1~{R},6~{R},8~{R},9~{R},10~{R},15~{R},17~{R},18~{R})-17-(2-azanyl-6-oxidanylidene-1~{H}-purin-9-yl)-9,18-bis(fluoranyl)-3,12-bis(oxidanyl)-3,12-bis(oxidanylidene)-2,4,7,11,13,16-hexaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.3.0.0^{6,10}]octadecan-8-yl]-1~{H}-purin-6-one, Stimulator of interferon protein | | Authors: | Boura, E, Smola, M. | | Deposit date: | 2020-03-20 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Ligand Strain and Its Conformational Complexity Is a Major Factor in the Binding of Cyclic Dinucleotides to STING Protein.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6ED9
 
 | |
6YDZ
 
 | | Human wtSTING in complex with 3',3'-cGAMP | | Descriptor: | 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, Stimulator of interferon protein | | Authors: | Boura, E, Smola, M. | | Deposit date: | 2020-03-23 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Ligand Strain and Its Conformational Complexity Is a Major Factor in the Binding of Cyclic Dinucleotides to STING Protein.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6YEA
 
 | | Human wtSTING in complex with 2',2'-difluoro-3',3'-cGAMP | | Descriptor: | 2',2'-difluoro-3',3'-cGAMP, Stimulator of interferon protein | | Authors: | Boura, E, Smola, M. | | Deposit date: | 2020-03-24 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | Ligand Strain and Its Conformational Complexity Is a Major Factor in the Binding of Cyclic Dinucleotides to STING Protein.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6ER2
 
 | |
6YHY
 
 | | A lid blocking mechanism of a cone snail toxin revealed at the atomic level | | Descriptor: | Conk-S1 | | Authors: | Saikia, C, Altman-Gueta, H, Dym, O, Frolow, F, Gurevitz, M, Gordon, D, Reuveny, E, Karbat, I. | | Deposit date: | 2020-03-31 | | Release date: | 2021-04-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A Molecular Lid Mechanism of K + Channel Blocker Action Revealed by a Cone Peptide.
J.Mol.Biol., 433, 2021
|
|
6YL2
 
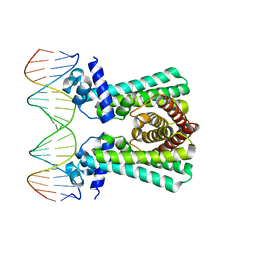 | | Structural and DNA binding studies of the transcriptional repressor Rv2506 (BkaR) from Mycobacterium tuberculosis supports a role in L-Leucine catabolism | | Descriptor: | DNA (5'-D(*AP*CP*GP*TP*TP*AP*AP*TP*GP*AP*CP*GP*AP*TP*TP*AP*AP*CP*CP*G)-3'), DNA (5'-D(*CP*GP*GP*TP*TP*AP*AP*TP*CP*GP*TP*CP*AP*TP*TP*AP*AP*CP*GP*T)-3'), Probable transcriptional regulatory protein (Probably TetR-family) | | Authors: | Keep, N.H, Pritchard, J.E, Sula, A, Cole, A.R, Kendall, S.L. | | Deposit date: | 2020-04-06 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural and DNA binding studies of the transcriptional repressor Rv2506 (BkaR) from Mycobacterium tuberculosis supports a role in L-Leucine catabolism
To be published
|
|
6EJA
 
 | |
6YBE
 
 | |
6YEL
 
 | | Stromal interaction molecule 1 coiled-coil 1 fragment | | Descriptor: | Stromal interaction molecule 1 | | Authors: | Rathner, P, Cerofolini, L, Ravera, E, Bechmann, M, Grabmayr, H, Fahrner, M, Fragai, M, Romanin, C, Luchinat, C, Mueller, N. | | Deposit date: | 2020-03-25 | | Release date: | 2020-09-02 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Interhelical interactions within the STIM1 CC1 domain modulate CRAC channel activation.
Nat.Chem.Biol., 17, 2021
|
|
6Z7X
 
 | | Insulin analytical antibody OXI-005 Fab | | Descriptor: | OXI-005 Fab Heavy chain, OXI-005 Fab Light chain | | Authors: | Johansson, E. | | Deposit date: | 2020-06-02 | | Release date: | 2020-12-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Insulin binding to the analytical antibody sandwich pair OXI-005 and HUI-018: Epitope mapping and binding properties.
Protein Sci., 30, 2021
|
|
6EZ6
 
 | | PI3 kinase delta in complex with Methyl 5-(4-(5-((4-isopropylpiperazin-1-yl)methyl)oxazol-2-yl)-1H-indazol-6-yl)-2-methoxynicotinate | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, methyl 2-methoxy-5-[4-[5-[(4-propan-2-ylpiperazin-1-yl)methyl]-1,3-oxazol-2-yl]-2~{H}-indazol-6-yl]pyridine-3-carboxylate | | Authors: | Convery, M.A, Campos, S, Dalton, S.E. | | Deposit date: | 2017-11-14 | | Release date: | 2017-12-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Selectively Targeting the Kinome-Conserved Lysine of PI3K delta as a General Approach to Covalent Kinase Inhibition.
J. Am. Chem. Soc., 140, 2018
|
|
6F0Q
 
 | |
6ZET
 
 | | Crystal structure of proteinase K nanocrystals by electron diffraction with a 20 micrometre C2 condenser aperture | | Descriptor: | CALCIUM ION, Proteinase K | | Authors: | Evans, G, Zhang, P, Beale, E.V, Waterman, D.G. | | Deposit date: | 2020-06-16 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.701 Å) | | Cite: | A Workflow for Protein Structure Determination From Thin Crystal Lamella by Micro-Electron Diffraction.
Front Mol Biosci, 7, 2020
|
|
