7TFJ
 
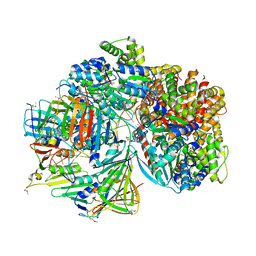 | | Atomic model of S. cerevisiae clamp-clamp loader complex PCNA-RFC bound to DNA with a closed clamp ring | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | 著者 | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | 登録日 | 2022-01-06 | | 公開日 | 2022-11-16 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Cryo-EM structures reveal that RFC recognizes both the 3'- and 5'-DNA ends to load PCNA onto gaps for DNA repair.
Elife, 11, 2022
|
|
4J3F
 
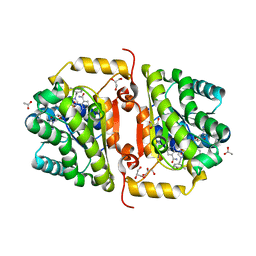 | | Crystal Structure of FabI from F. tularensis in complex with novel inhibitors based on the benzimidazole scaffold. | | 分子名称: | 1-(4-methoxy-3-methylbenzyl)-5,6,7,8-tetrahydro-1H-naphtho[2,3-d]imidazole, ACETATE ION, Enoyl-[acyl-carrier-protein] reductase [NADH], ... | | 著者 | Mehboob, S, Boci, T, Brubaker, L, Santarsiero, B.D, Johnson, M.E. | | 登録日 | 2013-02-05 | | 公開日 | 2014-07-23 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural and biological evaluation of a novel series of benzimidazole inhibitors of Francisella tularensis enoyl-ACP reductase (FabI).
Bioorg.Med.Chem.Lett., 25, 2015
|
|
2WNT
 
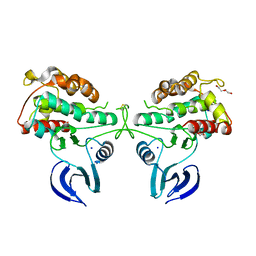 | | Crystal Structure of the Human Ribosomal protein S6 kinase | | 分子名称: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, RIBOSOMAL PROTEIN S6 KINASE, ... | | 著者 | Muniz, J.R.C, Elkins, J.M, Wang, J, Ugochukwu, E, Salah, E, King, O, Picaud, S, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Knapp, S. | | 登録日 | 2009-07-20 | | 公開日 | 2009-08-25 | | 最終更新日 | 2018-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal Structure of the Human Ribosomal Protein S6 Kinase
To be Published
|
|
4FK9
 
 | |
4QV6
 
 | | yCP beta5-A49V mutant | | 分子名称: | CHLORIDE ION, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | 著者 | Huber, E.M, Heinemeyer, W, Groll, M. | | 登録日 | 2014-07-14 | | 公開日 | 2015-02-04 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
3P5Z
 
 | | Crystal structure of the mutant T159S of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | 分子名称: | 6-HYDROXYURIDINE-5'-PHOSPHATE, GLYCEROL, N-PROPANOL, ... | | 著者 | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | 登録日 | 2010-10-11 | | 公開日 | 2011-09-21 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Conformational changes in orotidine 5'-monophosphate decarboxylase: a structure-based explanation for how the 5'-phosphate group activates the enzyme.
Biochemistry, 51, 2012
|
|
7U0H
 
 | | State NE1 nucleolar 60S ribosome biogenesis intermediate - Overall model | | 分子名称: | 25S rRNA, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, 5.8S rRNA, ... | | 著者 | Cruz, V.E, Sekulski, K, Peddada, N, Erzberger, J.P. | | 登録日 | 2022-02-18 | | 公開日 | 2022-12-14 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (2.76 Å) | | 主引用文献 | Sequence-specific remodeling of a topologically complex RNP substrate by Spb4.
Nat.Struct.Mol.Biol., 29, 2022
|
|
4QW7
 
 | | yCP beta5-M45T mutant in complex with carfilzomib | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | 著者 | Huber, E.M, Heinemeyer, W, Groll, M. | | 登録日 | 2014-07-16 | | 公開日 | 2015-02-04 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
4QXJ
 
 | | yCP beta5-M45A mutant in complex with the epoxyketone inhibitor ONX 0914 | | 分子名称: | 1,2,4-trideoxy-4-methyl-2-{[N-(morpholin-4-ylacetyl)-L-alanyl-O-methyl-L-tyrosyl]amino}-1-phenyl-D-xylitol, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | 著者 | Huber, E.M, Heinemeyer, W, Groll, M. | | 登録日 | 2014-07-21 | | 公開日 | 2015-02-04 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
4QZW
 
 | | yCP beta5-C52F mutant in complex with the epoxyketone inhibitor ONX 0914 | | 分子名称: | 1,2,4-trideoxy-4-methyl-2-{[N-(morpholin-4-ylacetyl)-L-alanyl-O-methyl-L-tyrosyl]amino}-1-phenyl-D-xylitol, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | 著者 | Huber, E.M, Heinemeyer, W, Groll, M. | | 登録日 | 2014-07-29 | | 公開日 | 2015-02-04 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
4A6Z
 
 | | Cytochrome c peroxidase with bound guaiacol | | 分子名称: | CYTOCHROME C PEROXIDASE, MITOCHONDRIAL, Guaiacol, ... | | 著者 | Murphy, E.J, Metcalfe, C.L, Nnamchi, C, Raven, E.L, Moody, P.C.E. | | 登録日 | 2011-11-10 | | 公開日 | 2012-10-17 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.61 Å) | | 主引用文献 | Crystal Structure of Guaiacol and Phenol Bound to a Heme Peroxidase.
FEBS J., 279, 2012
|
|
3LJG
 
 | | Human MMP12 in complex with non-zinc chelating inhibitor | | 分子名称: | ACETOHYDROXAMIC ACID, CALCIUM ION, HEXAETHYLENE GLYCOL, ... | | 著者 | Stura, E.A, Dive, V, Devel, L, Vera, L, Beau, F. | | 登録日 | 2010-01-26 | | 公開日 | 2010-09-01 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.313 Å) | | 主引用文献 | Insights from selective non-phosphinic inhibitors of MMP-12 tailored to fit with an S1' loop canonical conformation.
J.Biol.Chem., 285, 2010
|
|
1MXK
 
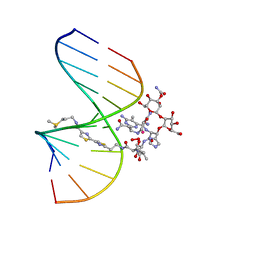 | | NMR Structure of HO2-Co(III)bleomycin A(2) Bound to d(GGAAGCTTCC)(2) | | 分子名称: | 5'-D(*GP*GP*AP*AP*GP*CP*TP*TP*CP*C)-3', BLEOMYCIN A2, COBALT (III) ION, ... | | 著者 | Zhao, C, Xia, C, Mao, Q, Forsterling, H, DeRose, E, Antholine, W.E, Subczynski, W.K, Petering, D.H. | | 登録日 | 2002-10-02 | | 公開日 | 2002-10-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structures of HO(2)-Co(III)bleomycin A(2) Bound to d(GAGCTC)(2) and d(GGAAGCTTCC)(2): Structure-Reactivity Relationships of Co and Fe Bleomycins
J.Inorg.Biochem., 91, 2002
|
|
5H22
 
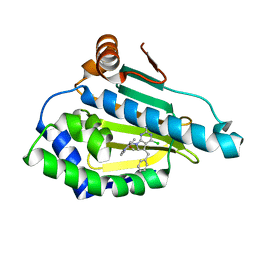 | | Hsp90 alpha N-terminal domain in complex with an inhibitor | | 分子名称: | 4-chloranyl-7-[(4-methoxy-3,5-dimethyl-pyridin-2-yl)methyl]-5-(phenylmethyl)pyrrolo[2,3-d]pyrimidin-2-amine, Hsp90aa1 protein | | 著者 | Kim, E.E, Shin, S.C, Keum, G.C. | | 登録日 | 2016-10-13 | | 公開日 | 2017-10-11 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.499 Å) | | 主引用文献 | Synthesis and in vitro antiproliferative activity of C5-benzyl substituted 2-amino-pyrrolo[2,3-d]pyrimidines as potent Hsp90 inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
4OL8
 
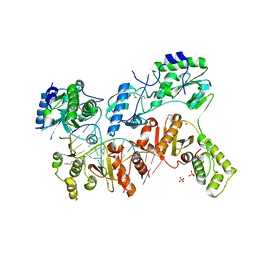 | | Ty3 reverse transcriptase bound to DNA/RNA | | 分子名称: | 5'-D(*CP*AP*TP*CP*TP*TP*CP*CP*TP*CP*TP*CP*TP*CP*TP*C)-3', 5'-R(*CP*UP*GP*AP*GP*AP*GP*AP*GP*AP*GP*GP*AP*AP*GP*AP*UP*G)-3', Reverse transcriptase/ribonuclease H, ... | | 著者 | Nowak, E, Miller, J.T, Bona, M.K, Studnicka, J, Szczepanowski, R.H, Jurkowski, J, Le Grice, S.F.J, Nowotny, M. | | 登録日 | 2014-01-23 | | 公開日 | 2014-03-05 | | 最終更新日 | 2014-04-16 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Ty3 reverse transcriptase complexed with an RNA-DNA hybrid shows structural and functional asymmetry.
Nat.Struct.Mol.Biol., 21, 2014
|
|
2WTI
 
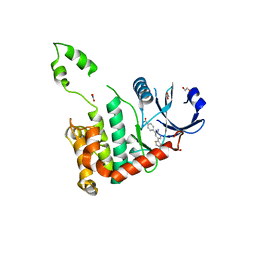 | | CRYSTAL STRUCTURE OF CHK2 IN COMPLEX WITH AN INHIBITOR | | 分子名称: | 1,2-ETHANEDIOL, 4-[2-AMINO-5-(2,3-DIHYDROTHIENO[3,4-B][1,4]DIOXIN-5-YL)PYRIDIN-3-YL]BENZAMIDE, CHECKPOINT KINASE 2, ... | | 著者 | Hilton, S, Naud, S, Caldwell, J.J, Boxall, K, Burns, S, Anderson, V.E, Antoni, L, Allen, C.E, Pearl, L.H, Oliver, A.W, Aherne, G.W, Garrett, M.D, Collins, I. | | 登録日 | 2009-09-16 | | 公開日 | 2009-12-29 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Identification and Characterisation of 2-Aminopyridine Inhibitors of Checkpoint Kinase 2
Bioorg.Med.Chem., 18, 2010
|
|
8QTC
 
 | |
8QM3
 
 | |
250L
 
 | | THE RESPONSE OF T4 LYSOZYME TO LARGE-TO-SMALL SUBSTITUTIONS WITHIN THE CORE AND ITS RELATION TO THE HYDROPHOBIC EFFECT | | 分子名称: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, T4 LYSOZYME | | 著者 | Xu, J, Baase, W.A, Baldwin, E, Matthews, B.W. | | 登録日 | 1997-10-22 | | 公開日 | 1998-03-18 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The response of T4 lysozyme to large-to-small substitutions within the core and its relation to the hydrophobic effect.
Protein Sci., 7, 1998
|
|
1QB5
 
 | |
4GEC
 
 | | Crystal Structure of E.coli MenH R124A Mutant | | 分子名称: | 1,2-ETHANEDIOL, 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase, CHLORIDE ION, ... | | 著者 | Johnston, J.M, Baker, E.N, Guo, Z, Jiang, M. | | 登録日 | 2012-08-01 | | 公開日 | 2013-05-08 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal Structures of E. coli Native MenH and Two Active Site Mutants.
Plos One, 8, 2013
|
|
8JHH
 
 | | Glycoside hydrolase family 55 endo-beta-1,3-glucanase from Microdochium nivale | | 分子名称: | GLYCEROL, MnLam55A | | 著者 | Ota, T, Saburi, W, Yamashita, K, Tagami, T, Yu, J, Komba, S, Jewell, L.E, Hsiang, T, Imai, R, Yao, M, Mori, H. | | 登録日 | 2023-05-23 | | 公開日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Molecular mechanism for endo-type action of glycoside hydrolase family 55 endo-beta-1,3-glucanase on beta 1-3/1-6-glucan.
J.Biol.Chem., 299, 2023
|
|
5CN7
 
 | | Ultrafast dynamics in myoglobin: 0.2 ps time delay | | 分子名称: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Barends, T.R.M, Foucar, L, Ardevol, A, Nass, K.J, Aquila, A, Botha, S, Doak, R.B, Falahati, K, Hartmann, E, Hilpert, M, Heinz, M, Hoffmann, M.C, Koefinger, J, Koglin, J, Kovacsova, G, Liang, M, Milathianaki, D, Lemke, H.T, Reinstein, J, Roome, C.M, Shoeman, R.L, Williams, G.J, Burghardt, I, Hummer, G, Boutet, S, Schlichting, I. | | 登録日 | 2015-07-17 | | 公開日 | 2015-09-16 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Direct observation of ultrafast collective motions in CO myoglobin upon ligand dissociation.
Science, 350, 2015
|
|
5CNE
 
 | | Ultrafast dynamics in myoglobin: 10 ps time delay | | 分子名称: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Barends, T.R.M, Foucar, L, Ardevol, A, Nass, K.J, Aquila, A, Botha, S, Doak, R.B, Falahati, K, Hartmann, E, Hilpert, M, Heinz, M, Hoffmann, M.C, Koefinger, J, Koglin, J, Kovacsova, G, Liang, M, Milathianaki, D, Lemke, H.T, Reinstein, J, Roome, C.M, Shoeman, R.L, Williams, G.J, Burghardt, I, Hummer, G, Boutet, S, Schlichting, I. | | 登録日 | 2015-07-17 | | 公開日 | 2015-09-16 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Direct observation of ultrafast collective motions in CO myoglobin upon ligand dissociation.
Science, 350, 2015
|
|
5KE3
 
 | | Crystal structure of SETDB1 Tudor domain in complex with fragment MRT0181a | | 分子名称: | (S)-N-(furan-2-ylmethyl)-1-(1,2,3,4-tetrahydroisoquinoline-3-carbonyl)piperidine-4-carboxamide, BETA-MERCAPTOETHANOL, Histone-lysine N-methyltransferase SETDB1, ... | | 著者 | Dong, A, Iqbal, A, Mader, P, Dobrovetsky, E, Ferreira de Freitas, R, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Schapira, M, Brown, P.J, Structural Genomics Consortium (SGC) | | 登録日 | 2016-06-09 | | 公開日 | 2016-08-03 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of SETDB1 Tudor domain in complex with fragment MRT0181a
to be published
|
|
