8F1V
 
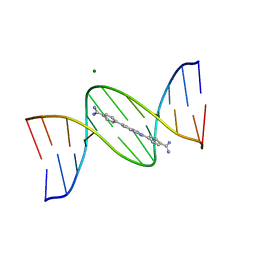 | | A benzimidazole (DB1476) sequence-specific recognition of 5'-CGCAAAAAAGCG-3' in B-orientation | | 分子名称: | 4,4'-(1H-benzimidazole-2,6-diyl)di(benzene-1-carboximidamide), DNA (5'-D(*CP*GP*CP*AP*AP*AP*AP*AP*AP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*TP*TP*TP*TP*TP*TP*GP*CP*G)-3'), ... | | 著者 | Ogbonna, E.N, Wilson, W.D. | | 登録日 | 2022-11-06 | | 公開日 | 2023-02-22 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | X-ray Structure Characterization of the Selective Recognition of AT Base Pair Sequences.
Acs Bio Med Chem Au, 3, 2023
|
|
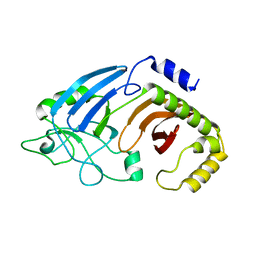
6XJ6
 
 | |
8F2Y
 
 | |
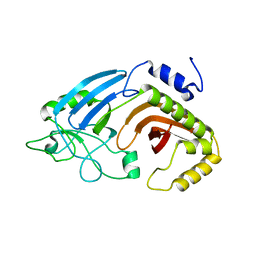
6XJ7
 
 | |
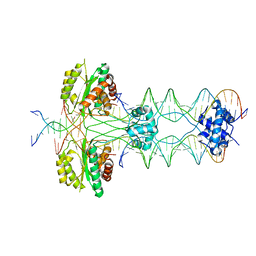
5HOO
 
 | |
7S6H
 
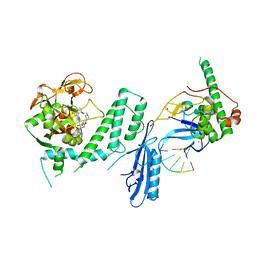 | | Human PARP1 deltaV687-E688 bound to NAD+ analog EB-47 and to a DNA double strand break. | | 分子名称: | 1,2-ETHANEDIOL, 2-[4-[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]carbonylpiperazin-1-yl]-N-(1-oxidanylidene-2,3-dihydroisoindol-4-yl)ethanamide, DNA (5'-D(*CP*GP*AP*CP*G)-3'), ... | | 著者 | Rouleau-Turcotte, E, Pascal, J.M. | | 登録日 | 2021-09-14 | | 公開日 | 2022-06-29 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Captured snapshots of PARP1 in the active state reveal the mechanics of PARP1 allostery.
Mol.Cell, 82, 2022
|
|
6EMY
 
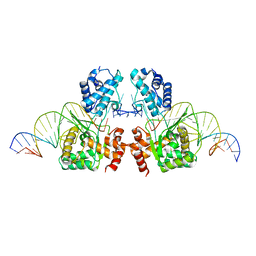 | | Structure of the Tn1549 transposon Integrase (aa 82-397, Y379F) in complex with transposon right end DNA | | 分子名称: | DNA (20-MER), DNA (26-MER), Int protein | | 著者 | Schulz, E.C, Rubio-Cosials, A, Barabas, O. | | 登録日 | 2017-10-04 | | 公開日 | 2018-04-04 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Transposase-DNA Complex Structures Reveal Mechanisms for Conjugative Transposition of Antibiotic Resistance.
Cell, 173, 2018
|
|
5LHB
 
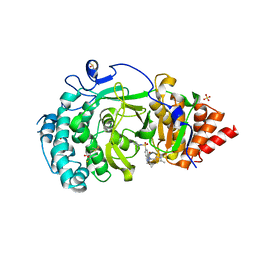 | | POLYADPRIBOSYL GLYCOSIDASE IN COMPLEX WITH PDD00017262 | | 分子名称: | 1-(cyclopropylmethyl)-6-[[(1-methylcyclopropyl)amino]-bis(oxidanyl)-$l^{4}-sulfanyl]-3-[(2-methyl-1,3-thiazol-5-yl)methyl]quinazoline-2,4-dione, DIMETHYL SULFOXIDE, Poly(ADP-ribose) glycohydrolase, ... | | 著者 | Tucker, J, Barkauskaite, E. | | 登録日 | 2016-07-10 | | 公開日 | 2016-10-12 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.23 Å) | | 主引用文献 | First-in-Class Chemical Probes against Poly(ADP-ribose) Glycohydrolase (PARG) Inhibit DNA Repair with Differential Pharmacology to Olaparib.
ACS Chem. Biol., 11, 2016
|
|
6XM9
 
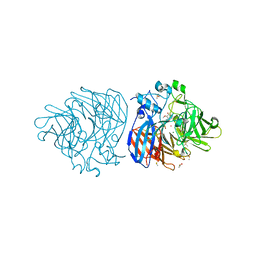 | | Crystal structure of vanillin bound to Co-LSD4 from Sphingobium sp. strain SYK-6 | | 分子名称: | 4-hydroxy-3-methoxybenzaldehyde, ACETATE ION, COBALT (II) ION, ... | | 著者 | Kuatsjah, E, Chan, A.C, Katahira, R, Beckham, G.T, Murphy, M.E, Eltis, L.D. | | 登録日 | 2020-06-29 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.651 Å) | | 主引用文献 | Structural and functional analysis of lignostilbene dioxygenases from Sphingobium sp. SYK-6.
J.Biol.Chem., 296, 2021
|
|
6OF1
 
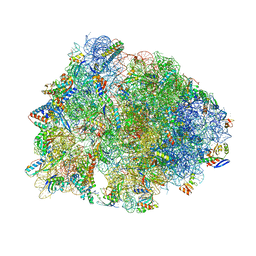 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with dirithromycin and bound to mRNA and A-, P-, and E-site tRNAs at 2.80A resolution | | 分子名称: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Khabibullina, N.F, Tereshchenkov, A.G, Komarova, E.S, Syroegin, E.A, Shiriaev, D.I, Paleskava, A, Kartsev, V.G, Bogdanov, A.A, Konevega, A.L, Dontsova, O.A, Sergiev, P.V, Osterman, I.A, Polikanov, Y.S. | | 登録日 | 2019-03-28 | | 公開日 | 2019-04-17 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure of Dirithromycin Bound to the Bacterial Ribosome Suggests New Ways for Rational Improvement of Macrolides.
Antimicrob.Agents Chemother., 63, 2019
|
|
6OAB
 
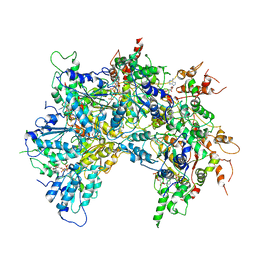 | | Cdc48-Npl4 complex processing poly-ubiquitinated substrate in the presence of ADP-BeFx, state 2 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Cell division control protein 48, ... | | 著者 | Twomey, E.C, Ji, Z, Wales, T.E, Bodnar, N.O, Engen, J.R, Rapoport, T.A. | | 登録日 | 2019-03-15 | | 公開日 | 2019-07-03 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Substrate processing by the Cdc48 ATPase complex is initiated by ubiquitin unfolding.
Science, 365, 2019
|
|
6XL4
 
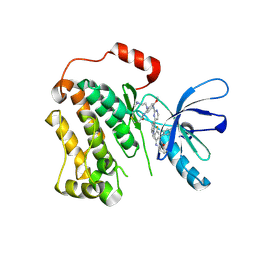 | | EGFR(T790M/V948R) in complex with AZD9291 and DDC4002 | | 分子名称: | 10-benzyl-8-fluoro-5,10-dihydro-11H-dibenzo[b,e][1,4]diazepin-11-one, Epidermal growth factor receptor, MAGNESIUM ION, ... | | 著者 | Heppner, D.E, Beyett, T.S, Eck, M.J. | | 登録日 | 2020-06-28 | | 公開日 | 2021-06-30 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | Molecular basis for cooperative binding and synergy of ATP-site and allosteric EGFR inhibitors.
Nat Commun, 13, 2022
|
|
8ERM
 
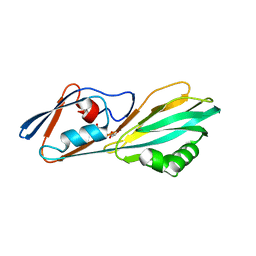 | | Crystal structure of FliC D2/D3 domains from Pseudomonas aeruginosa PAO1 | | 分子名称: | B-type flagellin, GLYCEROL, SULFATE ION | | 著者 | Nedeljkovic, M, Bonsor, D.A, Postel, S, Sundberg, E.J. | | 登録日 | 2022-10-12 | | 公開日 | 2023-05-17 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.475 Å) | | 主引用文献 | An unbroken network of interactions connecting flagellin domains is required for motility in viscous environments.
Plos Pathog., 19, 2023
|
|
8F0G
 
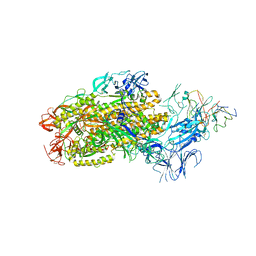 | | Structure of SARS-CoV-2 Omicron BA.1 spike in complex with antibody Fab 1C3 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody 1C3 Fab Heavy Chain, ... | | 著者 | Yu, X, Zyla, D, Hastie, K.M, Saphire, E.O. | | 登録日 | 2022-11-02 | | 公開日 | 2023-05-03 | | 実験手法 | ELECTRON MICROSCOPY (3.35 Å) | | 主引用文献 | Potent Omicron-neutralizing antibodies isolated from a patient vaccinated 6 months before Omicron emergence.
Cell Rep, 42, 2023
|
|
8F0H
 
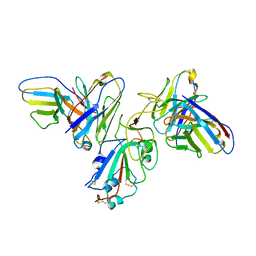 | | Structure of SARS-CoV-2 spike with antibody Fabs 2A10 and 1H2 (Local refinement of the RBD and Fabs 1H2 and 2A10) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody Fab 1H2 heavy chain, Antibody Fab 1H2 light chain, ... | | 著者 | Yu, X, Zyla, D, Hastie, K.M, Saphire, E.O. | | 登録日 | 2022-11-02 | | 公開日 | 2023-05-03 | | 実験手法 | ELECTRON MICROSCOPY (3.18 Å) | | 主引用文献 | Potent Omicron-neutralizing antibodies isolated from a patient vaccinated 6 months before Omicron emergence.
Cell Rep, 42, 2023
|
|
6ZZX
 
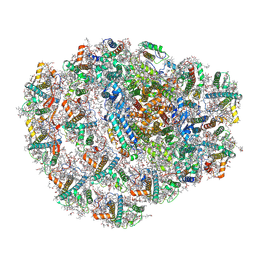 | | Structure of low-light grown Chlorella ohadii Photosystem I | | 分子名称: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (2S)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-hydroxypropyl hexadecanoate, (3R)-beta,beta-caroten-3-ol, ... | | 著者 | Caspy, I, Nelson, N, Nechushtai, R, Neumann, E, Shkolnisky, Y. | | 登録日 | 2020-08-05 | | 公開日 | 2021-07-28 | | 最終更新日 | 2021-09-29 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Cryo-EM photosystem I structure reveals adaptation mechanisms to extreme high light in Chlorella ohadii.
Nat.Plants, 7, 2021
|
|
7PUK
 
 | | Crystal structure of Endoglycosidase E GH18 domain from Enterococcus faecalis in complex with Man5 product | | 分子名称: | Beta-N-acetylhexosaminidase, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Garcia-Alija, M, Du, J.J, Trastoy, B, Sundberg, E.J, Guerin, M. | | 登録日 | 2021-09-30 | | 公開日 | 2022-03-16 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | Mechanism of cooperative N-glycan processing by the multi-modular endoglycosidase EndoE.
Nat Commun, 13, 2022
|
|
8F3H
 
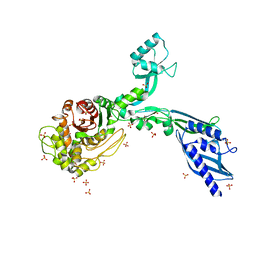 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) S466 insertion variant apo form from Enterococcus faecium | | 分子名称: | Penicillin binding protein 5, SULFATE ION | | 著者 | D'Andrea, E.D, Choy, M.S, Schoenle, M.V, Page, R, Peti, W. | | 登録日 | 2022-11-10 | | 公開日 | 2023-07-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F3L
 
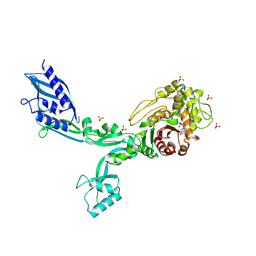 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) T485A variant penicillin bound form from Enterococcus faecium | | 分子名称: | OPEN FORM - PENICILLIN G, Penicillin binding protein 5, SULFATE ION | | 著者 | D'Andrea, E.D, Choy, M.S, Schoenle, M.V, Page, R, Peti, W. | | 登録日 | 2022-11-10 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F3O
 
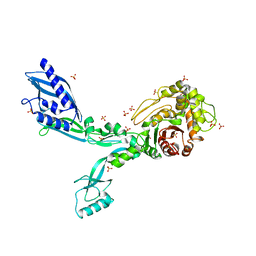 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) R464A variant apo form from Enterococcus faecium | | 分子名称: | Penicillin binding protein 5, SULFATE ION | | 著者 | D'Andrea, E.D, Choy, M.S, Schoenle, M.V, Peti, W, Page, R. | | 登録日 | 2022-11-10 | | 公開日 | 2023-07-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F3S
 
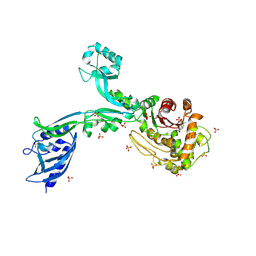 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) T485M T499I variant penicillin bound form from Enterococcus faecium | | 分子名称: | OPEN FORM - PENICILLIN G, Penicillin binding protein 5, SULFATE ION | | 著者 | D'Andrea, E.D, Choy, M.S, Schoenle, M.V, Page, R, Peti, W. | | 登録日 | 2022-11-10 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F67
 
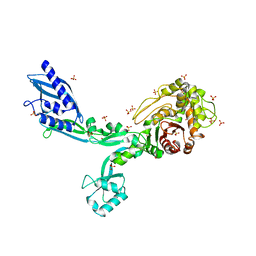 | | Crystal structure of the refolded Penicillin Binding Protein 5 (PBP5) of Enterococcus faecium | | 分子名称: | Pbp5, SULFATE ION | | 著者 | D'Andrea, E.D, Schoenle, M.V, Choy, M.S, Peti, W, Page, R. | | 登録日 | 2022-11-16 | | 公開日 | 2023-07-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.59 Å) | | 主引用文献 | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F3T
 
 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) T485M T499I V629E variant apo form from Enterococcus faecium | | 分子名称: | Penicillin binding protein 5, SODIUM ION, SULFATE ION | | 著者 | D'Andrea, E.D, Choy, M.S, Schoenle, M.V, Page, R, Peti, W. | | 登録日 | 2022-11-10 | | 公開日 | 2023-07-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.56 Å) | | 主引用文献 | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F3I
 
 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) S466 insertion variant penicillin bound form from Enterococcus faecium | | 分子名称: | OPEN FORM - PENICILLIN G, Penicillin binding protein 5, SULFATE ION | | 著者 | D'Andrea, E.D, Choy, M.S, Schoenle, M.V, Page, R, Peti, W. | | 登録日 | 2022-11-10 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F3U
 
 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) T485M T499I V629E variant penicillin bound form from Enterococcus faecium | | 分子名称: | OPEN FORM - PENICILLIN G, Penicillin binding protein 5, SULFATE ION | | 著者 | D'Andrea, E.D, Choy, M.S, Schoenle, M.V, Page, R, Peti, W. | | 登録日 | 2022-11-10 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
